|
Introduction
It should be noted that the PDB: 6OB0 was used to synthesize the following images.
(LPL) is an important enzyme for the breakdown of triglycerides in the body (Figure 1). [1] 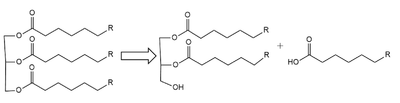 Figure 1: breakdown of a triglyceride into a diglyceride and creation of one free fatty acid by LPL A lipase is an enzyme that is capable of catalyzing the hydrolysis of fats/lipids which are consumed through oils. It is encoded by the p22 region in chromosome 8. Once synthesized, it is secreted into the interstitial space in several tissues. The main site of action for is in the capillary lumen within muscle and adipose tissues. [2] The function of this lipase is to hydrolyze triglycerides of very-low-density lipoproteins ( VLDL) and to aid in the delivery of lipid nutrients to vital tissues. [3] The enzyme is commonly found on the surface of cells that line blood capillaries. Two different lipoproteins are essential to break down triglycerides. One of the lipoproteins is utilized to transport fat into the bloodstream from different organs. [4] The lipoproteins essential, in the transport of fat from the intestine are referred to as chylomicrons. VLDL are utilized in carrying triglycerides from the liver into the bloodstream. The hydrolysis of triglycerides by lipoprotein lipase results in fat molecules being used by the body as energy or stored in fatty tissue. [5][6]
Structural Overview
LPL
is assumed to only be active as a composed of two LPL-GPIHBP1 heterodimers, however, previous studies have argued that the lipase can be active in its .[7][8]The N-terminal domain of lipoprotein lipase is known to consist of an alpha/beta hydrolase domain, which is composed of six alpha helices and ten beta strands. This domain creates an . The C-terminal domain of lipoprotein lipase is composed of twelve beta strands which form a "".[9]
GPIHBP1
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) is a secondary domain that is critical to the stabilization, function, and movement of .[10] The GPIHBP1’s highly acidic and intrinsically disordered N-terminal domain are essential to the binding of LPL’s C-terminal Domain. It has been shown that GPIHBP1 has a “three-fingered domain”, which holds it tightly to LPL by .[11] The importance of GPIBP1’s affinity to LPL was analyzed by Birrane et al.[12], and it was found that missense mutations of critical residues resulted in high amounts of impairments. It was also concluded that these impairments caused hypertriglyceridemia (chylomicronemia).[13]
Structural Highlights
Calcium Ion Stabilization
Ions are widely used in proteins and mechanistic stabilization in many areas of biochemistry. LPL’s tertiary folding is stabilized by a Calcium (Ca2+) ion. The calcium ion shares electron density with surrounding residues in order to orient the protein in its formal state.[14] The is achieved by the calcium ion’s interactions with the following of LPL’s residues: Ala194, Arg197, Ser199, Asp201, and Asp202.[15]
Lid and Lipid Binding Region
In the presence of the GPIHBP1 inhibitor, the and become visible within the structure. As displayed through a study conducted by Arora et. al, in 2019, the lipid-binding region of LPL actively interacts with the known inhibitor in the heterodimeric form. [16] This was established to be the only time that the heterodimeric form was shown as an active lipase. The lid region residues Ile245, Ile249, V251, Ile252, Leu257, Val260, Leu263, and Val264, are found as an open conformation which is composed of two small alpha helices that reach out and away from the protein. The lid and lipid-binding region create hydrophobic patches on the surface of lipoprotein lipase which are essential for by LPL.
Mechanism
Lipoprotein Lipase functions to catalyze the hydrolysis of one ester bond of triglycerides in order to remove one fatty acid tail and turn the triglyceride into a diglyceride. It does this by utilizing a simple serine hydrolase mechanism, in which it uses a composed of Asp183, His268, and Ser159 to catalyze the hydrolysis. His268 serves as a base catalyst by deprotonation of Ser159, which can then serve as the nucleophile to attack the carbonyl carbon of one of the fatty acid chains of a triglyceride. This forms a tetrahedral intermediate, which is stabilized by the amide of Trp82 and Leu160 residues, called the . The single fatty acid chain is cleaved from the triglyceride, forming the diglyceride product. Then, water is used to free the fatty acid from Ser159 and return all of the residues back to their starting point, in order to perform the mechanism again on another triglyceride. Ultimately, the hydrolysis results in the formation of one free fatty acid and glycerol with two fatty acid tails (Figure 2).
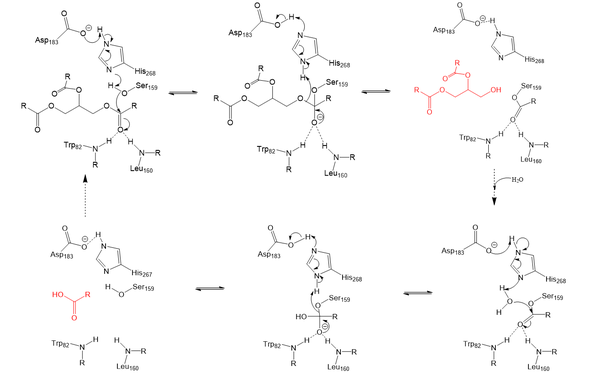 Figure 2: Serine hydrolase mechanism utilized by LPL to catalyze the breakdown of one ester bond of a triglyceride. Compounds colored red are the products of the hydrolysis. Relevance & Disease
LPL is an extremely important enzyme, in that it is responsible for the proper breakdown of certain fats in the body. LPL breaks down triglycerides carried in chylomicrons, otherwise known as very-low-density lipoproteins (VLDL). Chylomicrons carry digested fats in the form of triglycerides out of the small intestine and into the bloodstream. LPL recognizes the chylomicrons, and hydrolyzes the associated triglycerides.[17] When triglycerides are not broken down properly and are allowed to build up, they can lead to increased plasma triglyceride levels (hypertriglyceridemia) and cholesterol buildup. Hypertriglyceridemia is very unhealthy and is the leading cause of Coronary Artery Disease in America.[18] Cholesterol buildup is caused by excess fats (triglycerides) and is a similarly serious issue with regards to obesity and heart disease in the United States as it can lead to plaque buildup in arteries and veins, which restricts blood flow.[19] Chylomicronemia, which is defined as an excess of chylomicrons in the blood, is the disease characterized by the body being deficient in LPL resulting in persistent hypertriglyceridemia. This disease causes the body to be unable to digest very much ingested fats and often leads to severe abdominal discomfort and several episodes of acute pancreatitis.[20] In short, without LPL in the body, triglycerides are unable to get broken down, and there is a much higher likelihood of developing coronary & metabolic based diseases.
M404 Mutation
A 2018 study [21] argued that mutations that inhibit the proper binding of GPIHBP1 and LPL are suspected to cause chylomicronemia. A mutation that is believed to cause chylomicronemia is known as . Within this mutation, the methionine is replaced with a large and hydrophilic arginine. While this missense mutation does not impact LPL secretion, it does affect the formation of the LPL-GPIHBP1 complex by disrupting LPL’s interaction with GPIHB1’s Val121, Glu122, Thr124, and Val126 residues. This disrupts the stabilization of the LPL-GPIHBP1 complex, which has been seen to negatively affect LPL’s ability to catalyze the hydrolysis of triglycerides.
References
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med (Berl). 2002 Dec;80(12):753-69. doi: 10.1007/s00109-002-0384-9. Epub, 2002 Oct 24. PMID:12483461 doi:http://dx.doi.org/10.1007/s00109-002-0384-9
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Beigneux AP, Allan CM, Sandoval NP, Cho GW, Heizer PJ, Jung RS, Stanhope KL, Havel PJ, Birrane G, Meiyappan M, Gill JE 4th, Murakami M, Miyashita K, Nakajima K, Ploug M, Fong LG, Young SG. Lipoprotein lipase is active as a monomer. Proc Natl Acad Sci U S A. 2019 Mar 26;116(13):6319-6328. doi:, 10.1073/pnas.1900983116. Epub 2019 Mar 8. PMID:30850549 doi:http://dx.doi.org/10.1073/pnas.1900983116
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ Kersten S. Physiological regulation of lipoprotein lipase. Biochim Biophys Acta. 2014 Jul;1841(7):919-33. doi: 10.1016/j.bbalip.2014.03.013., Epub 2014 Apr 8. PMID:24721265 doi:http://dx.doi.org/10.1016/j.bbalip.2014.03.013
- ↑ Austin MA, Hokanson JE, Edwards KL. Hypertriglyceridemia as a cardiovascular risk factor. Am J Cardiol. 1998 Feb 26;81(4A):7B-12B. doi: 10.1016/s0002-9149(98)00031-9. PMID:9526807 doi:http://dx.doi.org/10.1016/s0002-9149(98)00031-9
- ↑ Kruth HS. Lipoprotein cholesterol and atherosclerosis. Curr Mol Med. 2001 Dec;1(6):633-53. doi: 10.2174/1566524013363212. PMID:11899253 doi:http://dx.doi.org/10.2174/1566524013363212
- ↑ Francis A, Levy Y. [Chylomicronemia syndrome]. Harefuah. 2002 Feb;141(2):201-3, 221, 220. PMID:11905095
- ↑ Paquette M, Hegele RA, Pare G, Baass A. A novel mutation in GPIHBP1 causes familial chylomicronemia syndrome. J Clin Lipidol. 2018 Mar - Apr;12(2):506-510. doi: 10.1016/j.jacl.2018.01.011., Epub 2018 Jan 31. PMID:29452893 doi:http://dx.doi.org/10.1016/j.jacl.2018.01.011
Student Contributors
Giselle Flores
Dustin Soe
Maggie Stopa
|


