This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
2pol
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
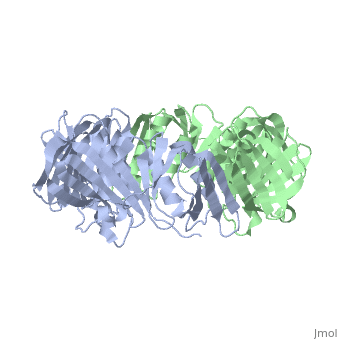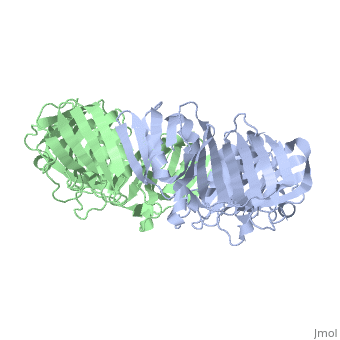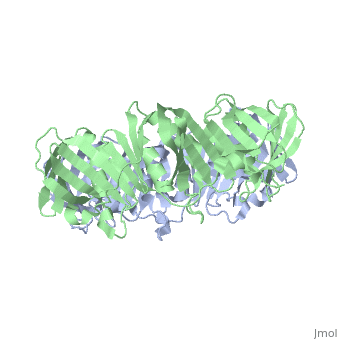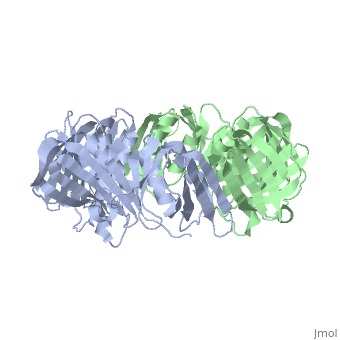
==THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP== | ==THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP== | ||
| - | <StructureSection load='2pol' size='340' side='right' caption='[[2pol]], [[Resolution|resolution]] 2.50Å' scene=''> | + | <StructureSection load='2pol' size='340' side='right'caption='[[2pol]], [[Resolution|resolution]] 2.50Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2pol]] is a 2 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[2pol]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/"bacillus_coli"_migula_1895 "bacillus coli" migula 1895]. The June 2012 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Sliding Clamps'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2012_6 10.2210/rcsb_pdb/mom_2012_6]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2POL OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2POL FirstGlance]. <br> |
| - | </td></tr><tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">POL ([ | + | </td></tr><tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">POL ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 "Bacillus coli" Migula 1895])</td></tr> |
| - | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[https://en.wikipedia.org/wiki/DNA-directed_DNA_polymerase DNA-directed DNA polymerase], with EC number [https://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.7.7 2.7.7.7] </span></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2pol FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pol OCA], [https://pdbe.org/2pol PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2pol RCSB], [https://www.ebi.ac.uk/pdbsum/2pol PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2pol ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/DPO3B_ECOLI DPO3B_ECOLI]] DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity. The beta chain is required for initiation of replication once it is clamped onto DNA, it slides freely (bidirectional and ATP-independent) along duplex DNA. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 29: | Line 29: | ||
</div> | </div> | ||
<div class="pdbe-citations 2pol" style="background-color:#fffaf0;"></div> | <div class="pdbe-citations 2pol" style="background-color:#fffaf0;"></div> | ||
| - | |||
| - | ==See Also== | ||
| - | *[[DNA polymerase|DNA polymerase]] | ||
| - | *[[Polymerase III homoenzyme beta subunit|Polymerase III homoenzyme beta subunit]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| Line 39: | Line 35: | ||
[[Category: Bacillus coli migula 1895]] | [[Category: Bacillus coli migula 1895]] | ||
[[Category: DNA-directed DNA polymerase]] | [[Category: DNA-directed DNA polymerase]] | ||
| + | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Sliding Clamps]] | [[Category: Sliding Clamps]] | ||
Revision as of 15:29, 17 June 2021
THREE-DIMENSIONAL STRUCTURE OF THE BETA SUBUNIT OF ESCHERICHIA COLI DNA POLYMERASE III HOLOENZYME: A SLIDING DNA CLAMP
| |||||||||||