We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1oap
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1oap' size='340' side='right'caption='[[1oap]], [[Resolution|resolution]] 1.93Å' scene=''> | <StructureSection load='1oap' size='340' side='right'caption='[[1oap]], [[Resolution|resolution]] 1.93Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
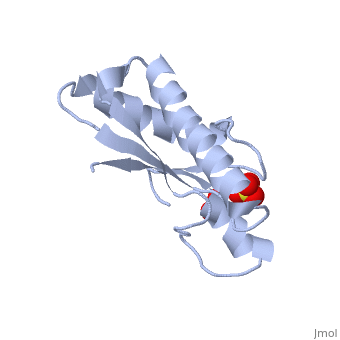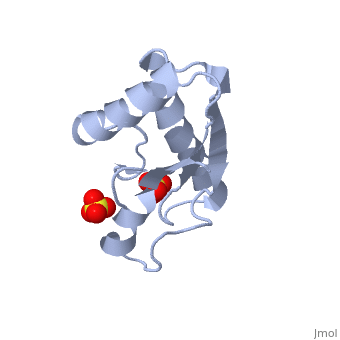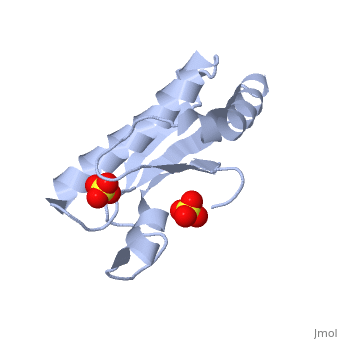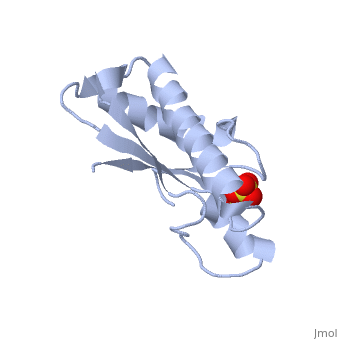
| - | <table><tr><td colspan='2'>[[1oap]] is a 1 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1oap]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/"bacillus_coli"_migula_1895 "bacillus coli" migula 1895]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1OAP OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1OAP FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1oap FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1oap OCA], [https://pdbe.org/1oap PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1oap RCSB], [https://www.ebi.ac.uk/pdbsum/1oap PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1oap ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/PAL_ECOLI PAL_ECOLI]] Thought to play a role in bacterial envelope integrity. Very strongly associated with the peptidoglycan. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Revision as of 09:51, 21 July 2021
Mad structure of the periplasmique domain of the Escherichia coli PAL protein
| |||||||||||