This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
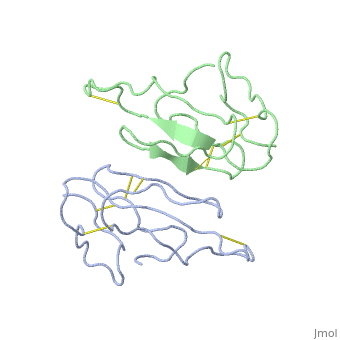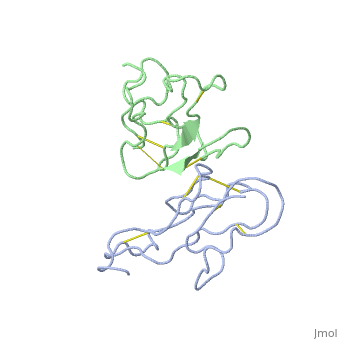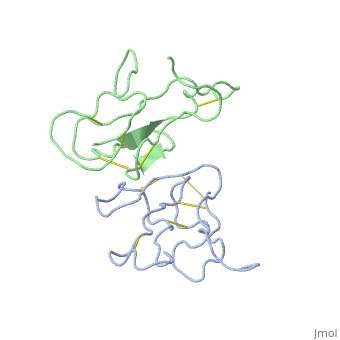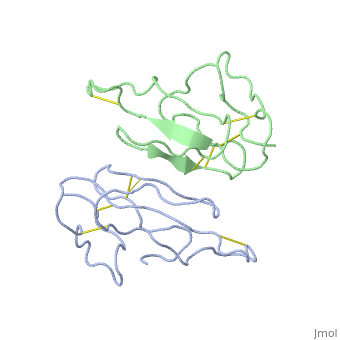
2abx
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='2abx' size='340' side='right'caption='[[2abx]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='2abx' size='340' side='right'caption='[[2abx]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2abx]] is a 2 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[2abx]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Bunmu Bunmu]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1abx 1abx]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2ABX OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2ABX FirstGlance]. <br> |
| - | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2abx FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2abx OCA], [https://pdbe.org/2abx PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2abx RCSB], [https://www.ebi.ac.uk/pdbsum/2abx PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2abx ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/NXL1A_BUNMU NXL1A_BUNMU]] Binds with high affinity to muscular and neuronal (alpha-7, alpha-8, and alpha-9) nicotinic acetylcholine receptors. Produces peripheral paralysis by blocking neuromuscular transmission at the postsynaptic site. Blocks the extracellular increase of dopamine evoked by nicotine only at the higher dose (4.2 uM).<ref>PMID:9305882</ref> <ref>PMID:9840221</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Revision as of 14:38, 17 November 2021
THE CRYSTAL STRUCTURE OF ALPHA-BUNGAROTOXIN AT 2.5 ANGSTROMS RESOLUTION. RELATION TO SOLUTION STRUCTURE AND BINDING TO ACETYLCHOLINE RECEPTOR
| |||||||||||