Transmembrane protease serine 2
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Protease activity == | == Protease activity == | ||
| - | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. This three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> | + | '''TMPRSS2''', as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. This three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> |
The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | ||
Revision as of 15:54, 29 November 2021
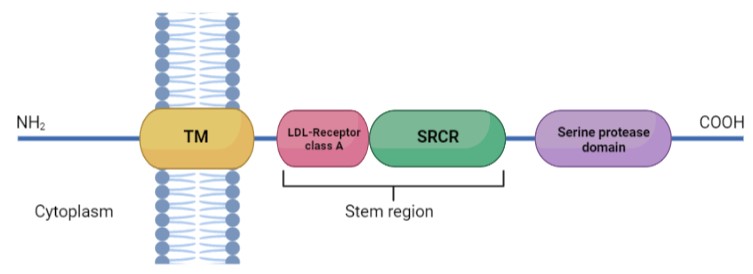
TMPRSS2 is a membrane protein belonging to the type II transmembrane serine protease (TTSP) family. It is functionally classified as a trypsin-like protease (TLP). [1] Serine proteases are known to be involved in many physiological and pathological processes.
| |||||||||||
References
- ↑ Sgrignani J, Cavalli A. Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2. Front Mol Biosci. 2021 Apr 30;8:666626. doi: 10.3389/fmolb.2021.666626., eCollection 2021. PMID:33996911 doi:http://dx.doi.org/10.3389/fmolb.2021.666626
- ↑ Evnin LB, Vasquez JR, Craik CS. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6659-63. doi: 10.1073/pnas.87.17.6659. PMID:2204062 doi:http://dx.doi.org/10.1073/pnas.87.17.6659
- ↑ Singh N, Decroly E, Khatib AM, Villoutreix BO. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur J Pharm Sci. 2020 Oct 1;153:105495. doi: 10.1016/j.ejps.2020.105495. Epub, 2020 Jul 28. PMID:32730844 doi:http://dx.doi.org/10.1016/j.ejps.2020.105495
- ↑ Lam DK, Dang D, Flynn AN, Hardt M, Schmidt BL. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain. 2015 May;156(5):923-930. doi: 10.1097/j.pain.0000000000000130. PMID:25734995 doi:http://dx.doi.org/10.1097/j.pain.0000000000000130
- ↑ Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. 2020 Dec;73(12):773-776. doi: 10.1136/jclinpath-2020-206987. Epub , 2020 Sep 1. PMID:32873700 doi:http://dx.doi.org/10.1136/jclinpath-2020-206987
Proteopedia Page Contributors and Editors (what is this?)
Paula R. Mallavibarrena, Ines Muniesa-Martinez, Michal Harel, Laura Aleixos Juan, Jaime Prilusky