User:Natalia de Araujo./Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
---- | ---- | ||
| - | The first structure | + | ''The first structure'', displayed on the left-hand side of the screen, corresponds to [[4jon]], the Crystal structure of CEP170's transcript variant beta, from Homo sapiens. 4jon is a 5 chain structure obtained by ''X-ray Crystallography'' [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions.&text=In%20a%20single%2Dcrystal%20X,is%20mounted%20on%20a%20goniometer.] and represents the first 126 residues of CEP170's N-termini region. |
| - | The second structure available for CEP170 is <scene name='89/897701/Q5sw79/1'>Q5SW79</scene>, a predicted 3D model. | + | '''The second structure''' available for CEP170 is <scene name='89/897701/Q5sw79/1'>Q5SW79</scene>, a ''predicted'' 3D model. |
Here we are going to focus on the '''Q5SW79''' model due to its full sequence, allowing us to highlight residues and regions important for the understanding of CEP170's functions and interactions. | Here we are going to focus on the '''Q5SW79''' model due to its full sequence, allowing us to highlight residues and regions important for the understanding of CEP170's functions and interactions. | ||
| Line 23: | Line 23: | ||
</table> | </table> | ||
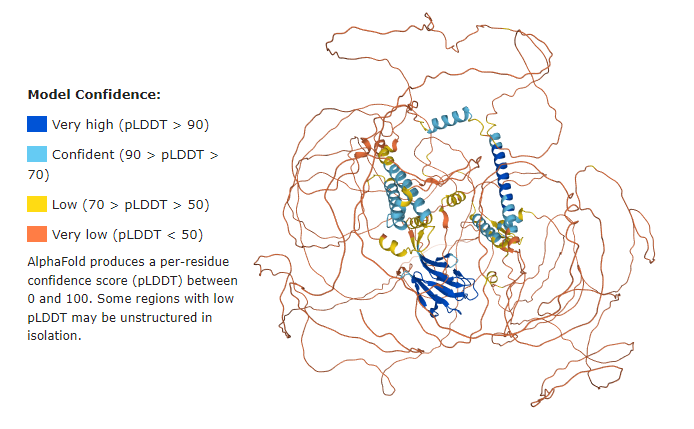
| - | == | + | == Q5SW79 Confidence Model == |
| - | + | Before getting started, it is important to have in mind that the majority of the Q5SW79 structure has {{Font color|orange|very low confidence}}, meaning the model most probably does not represent the real protein structure. | |
However, it stills helps us to have a good idea of the whole protein. | However, it stills helps us to have a good idea of the whole protein. | ||
| + | |||
| + | See below the confidence model from UniProt: | ||
<imagemap> | <imagemap> | ||
Revision as of 19:54, 3 December 2021
Centrosomal protein of 170kDa (CEP170)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo M, Wei R, Wang Y, Jurczyszyn A, Janz S, Beksac M, Zhan F, Seckinger A, Hose D, Pan J, Yang Y. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021 Jun 5;20(1):84. doi: 10.1186/s12943-021-01380-0. PMID:34090465 doi:http://dx.doi.org/10.1186/s12943-021-01380-0