2dda
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
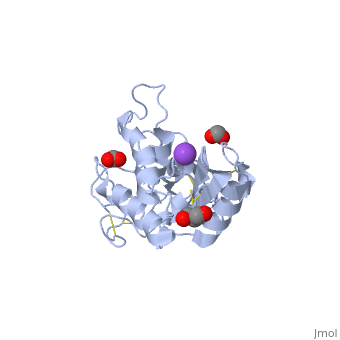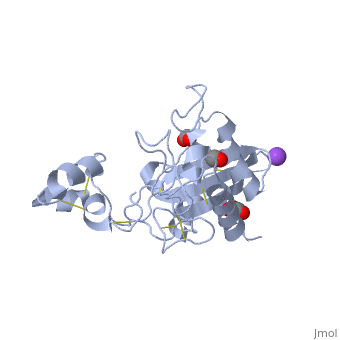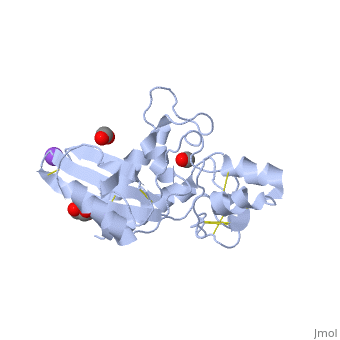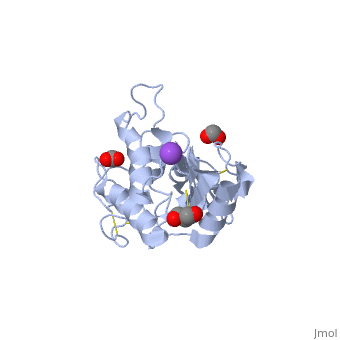
<StructureSection load='2dda' size='340' side='right'caption='[[2dda]], [[Resolution|resolution]] 2.25Å' scene=''> | <StructureSection load='2dda' size='340' side='right'caption='[[2dda]], [[Resolution|resolution]] 2.25Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2dda]] is a 4 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[2dda]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Pseudechis_australis Pseudechis australis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2DDA OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2DDA FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=FMT:FORMIC+ACID'>FMT</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=NA:SODIUM+ION'>NA</scene></td></tr> | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=FMT:FORMIC+ACID'>FMT</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=NA:SODIUM+ION'>NA</scene></td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[2ddb|2ddb]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[2ddb|2ddb]]</div></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2dda FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2dda OCA], [https://pdbe.org/2dda PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2dda RCSB], [https://www.ebi.ac.uk/pdbsum/2dda PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2dda ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/CRVP_PSEAU CRVP_PSEAU]] Blocks olfactory (CNGA2) and retinal (CNGA1) cyclic nucleotide-gated (CNG) ion channel currents. Does not inhibit retinal (CNGA3) currents. It forms high-affinity contacts with the pore turret region and most likely inhibits CNG channel current by blocking the external entrance to the transmembrane pore. Is really more potent that Pseudecin. Does not affect neither depolarization- nor caffeine-induced contraction arterial smooth muscle.<ref>PMID:12234174</ref> <ref>PMID:9892706</ref> <ref>PMID:14638933</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
Revision as of 10:30, 8 December 2021
Crystal structure of pseudechetoxin from Pseudechis australis
| |||||||||||
Categories: Large Structures | Pseudechis australis | Fujimoto, Z | Mizuno, H | Morita, T | Suzuki, N | Yamazaki, Y | Cng channel | Crisp | Snake venom | Toxin