User:Guilherme Gonzalez/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
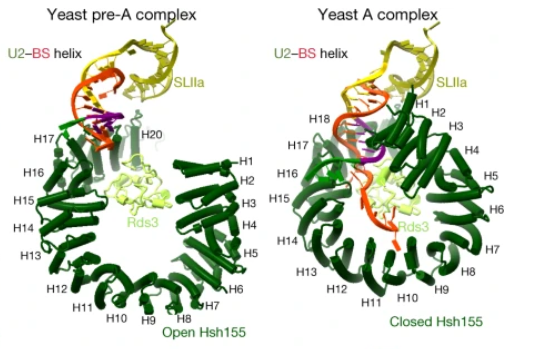
This structure is essential to the function of this protein, allowing ligants to interact with the innermost part of this molecule and alter its opening, being capable of small regulations in its capacity to interact with the pre-RNAm and other molecules of the U2 snRNA. Besides that, this structure has two main states: Open and Closed. This can be regulated and permits the interaction and the fixation of the pre-RNAm, and this process allows the formation of the A complex during the splicing, the closing of the HSH155<sup>HEAT</sup> that permits the assemble of the A complex<ref>DOI 10.1038/s41586-021-03789-5</ref>. | This structure is essential to the function of this protein, allowing ligants to interact with the innermost part of this molecule and alter its opening, being capable of small regulations in its capacity to interact with the pre-RNAm and other molecules of the U2 snRNA. Besides that, this structure has two main states: Open and Closed. This can be regulated and permits the interaction and the fixation of the pre-RNAm, and this process allows the formation of the A complex during the splicing, the closing of the HSH155<sup>HEAT</sup> that permits the assemble of the A complex<ref>DOI 10.1038/s41586-021-03789-5</ref>. | ||
| + | |||
| + | [[Image:HSHOpenClosed.png]] | ||
In the box at the right, it is possible to see its <scene name='89/897726/Chain_a__filledy/2'>general structure</scene> in a space-fill model obtained by electron microscopy at 9 Å resolution. However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='89/897726/Chain_a__secondaryy/1'>secondary structure patterns</scene>. | In the box at the right, it is possible to see its <scene name='89/897726/Chain_a__filledy/2'>general structure</scene> in a space-fill model obtained by electron microscopy at 9 Å resolution. However, to cover some important aspects of the structure and function of the molecule, it is particularly useful to represent its <scene name='89/897726/Chain_a__secondaryy/1'>secondary structure patterns</scene>. | ||
| Line 19: | Line 21: | ||
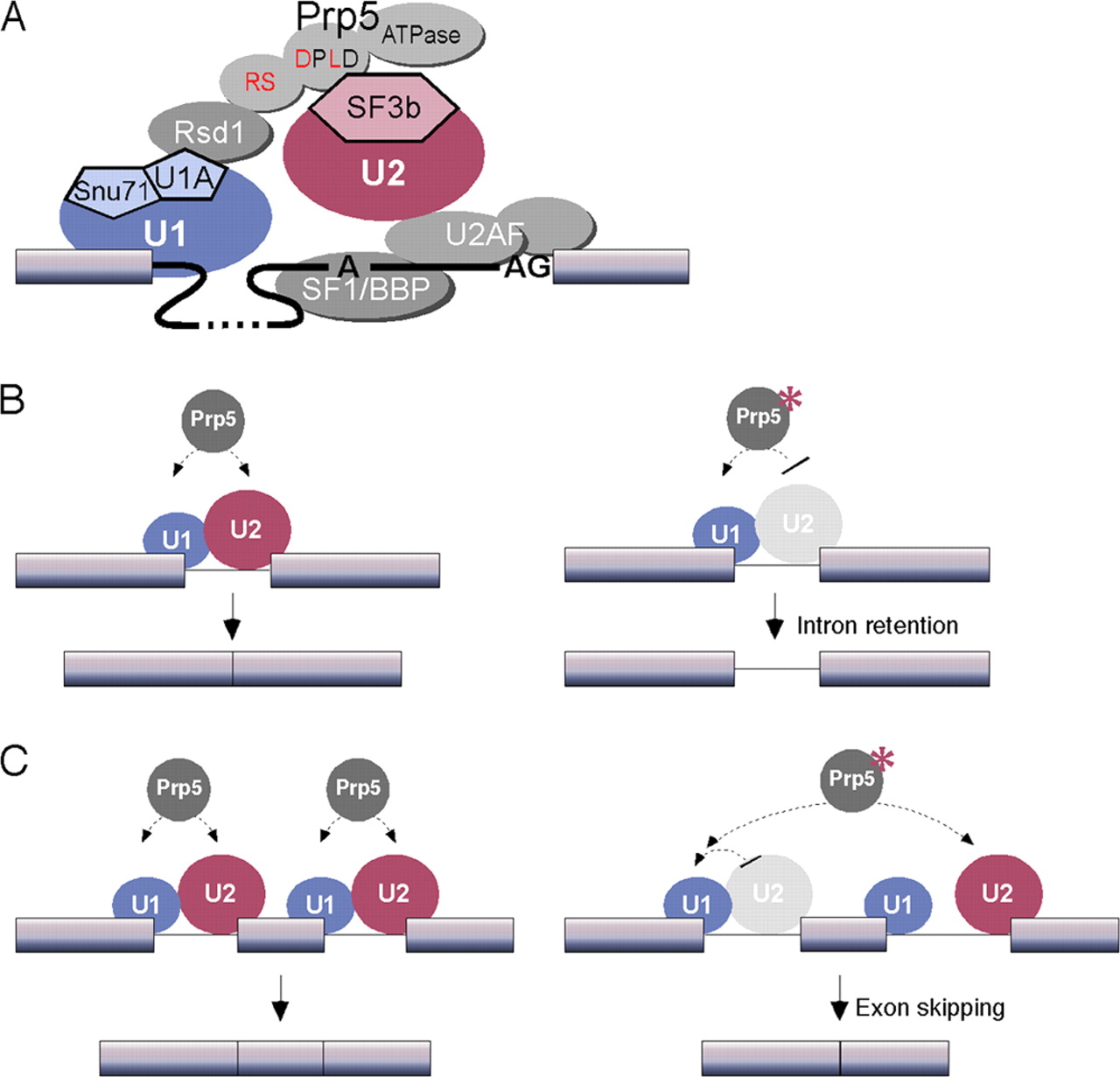
In eukaryotes the U2 snRNA is essential for the assembly of the Pre-A complex, because of the ability to recognize and bind to the intron on mRNA primary transcripts. This highly dependent on the HSH155<sup>HEAT</sup>, but the U2 snRNA also has been proposed to have a catalytic purpose on the splicing. Although this function is not a direct result of the interaction of the RNAm with the HSH155<sup>HEAT</sup>, this protein is also involved by regulating the attachment and the movement of associated proteins in this complex when it shifts between the open and close states<ref>DOI 10.1038/nature12734</ref>. | In eukaryotes the U2 snRNA is essential for the assembly of the Pre-A complex, because of the ability to recognize and bind to the intron on mRNA primary transcripts. This highly dependent on the HSH155<sup>HEAT</sup>, but the U2 snRNA also has been proposed to have a catalytic purpose on the splicing. Although this function is not a direct result of the interaction of the RNAm with the HSH155<sup>HEAT</sup>, this protein is also involved by regulating the attachment and the movement of associated proteins in this complex when it shifts between the open and close states<ref>DOI 10.1038/nature12734</ref>. | ||
| + | |||
| + | [[Image:U2U1 Splicing.jpeg]] | ||
The U2 snRNA can vary vastly between all eukaryotes, having different lengths and sequences according to the species. But that is also some very conservated parts of this complex and the HEAT repeats are one of them. This fact indicates how important this protein and this component are to the process of splicing<ref>10.1146/annurev.ge.22.120188.002131</ref>. | The U2 snRNA can vary vastly between all eukaryotes, having different lengths and sequences according to the species. But that is also some very conservated parts of this complex and the HEAT repeats are one of them. This fact indicates how important this protein and this component are to the process of splicing<ref>10.1146/annurev.ge.22.120188.002131</ref>. | ||
| Line 25: | Line 29: | ||
While the U2 snRNA identifies and binds with the branching point of the intron, the U1 snRNA identifies and binds with the 5’ splicing site. And together these two form the spliceosome A complex. This is one of the first steps needed to begin the process of splicing in eukaryotic cells<ref>DOI 10.1126/science.aaf4465</ref>. | While the U2 snRNA identifies and binds with the branching point of the intron, the U1 snRNA identifies and binds with the 5’ splicing site. And together these two form the spliceosome A complex. This is one of the first steps needed to begin the process of splicing in eukaryotic cells<ref>DOI 10.1126/science.aaf4465</ref>. | ||
| + | |||
Revision as of 15:00, 13 December 2021
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Mathew V, Tam AS, Milbury KL, Hofmann AK, Hughes CS, Morin GB, Loewen CJR, Stirling PC. Selective aggregation of the splicing factor Hsh155 suppresses splicing upon genotoxic stress. J Cell Biol. 2017 Dec 4;216(12):4027-4040. doi: 10.1083/jcb.201612018. Epub 2017 , Oct 4. PMID:28978642 doi:http://dx.doi.org/10.1083/jcb.201612018
- ↑ doi: https://dx.doi.org/10.2210/pdb7OQB/pdb
- ↑ Zhang Z, Rigo N, Dybkov O, Fourmann JB, Will CL, Kumar V, Urlaub H, Stark H, Luhrmann R. Structural insights into how Prp5 proofreads the pre-mRNA branch site. Nature. 2021 Aug;596(7871):296-300. doi: 10.1038/s41586-021-03789-5. Epub 2021, Aug 4. PMID:34349264 doi:http://dx.doi.org/10.1038/s41586-021-03789-5
- ↑ Zhan X, Yan C, Zhang X, Lei J, Shi Y. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res. 2018 Oct 12. pii: 10.1038/s41422-018-0094-7. doi:, 10.1038/s41422-018-0094-7. PMID:30315277 doi:http://dx.doi.org/10.1038/s41422-018-0094-7
- ↑ Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013 Nov 14;503(7475):229-34. doi: 10.1038/nature12734. Epub 2013 Nov 6. PMID:24196718 doi:http://dx.doi.org/10.1038/nature12734
- ↑ 10.1146/annurev.ge.22.120188.002131
- ↑ Cate JH. STRUCTURE. A Big Bang in spliceosome structural biology. Science. 2016 Mar 25;351(6280):1390-2. doi: 10.1126/science.aaf4465. PMID:27013712 doi:http://dx.doi.org/10.1126/science.aaf4465
- ↑ Malcovati L, Papaemmanuil E, Bowen DT, Boultwood J, Della Porta MG, Pascutto C, Travaglino E, Groves MJ, Godfrey AL, Ambaglio I, Galli A, Da Via MC, Conte S, Tauro S, Keenan N, Hyslop A, Hinton J, Mudie LJ, Wainscoat JS, Futreal PA, Stratton MR, Campbell PJ, Hellstrom-Lindberg E, Cazzola M. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011 Dec 8;118(24):6239-46. doi: 10.1182/blood-2011-09-377275. Epub 2011, Oct 12. PMID:21998214 doi:http://dx.doi.org/10.1182/blood-2011-09-377275