Journal:Acta Cryst F:S2053230X21012632
From Proteopedia
(Difference between revisions)

| Line 4: | Line 4: | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
| - | The high-resolution structure of a putative short-chain reductase from the commercially important bacteria ''Paraburkholderia xenovorans'' is reported (''Px''SDR). The reported apo structure of ''Px''SDR was determined in the monoclinic space group ''P''2<sub>1</sub> as a <scene name='89/899477/Cv/2'>prototypical SDR tetramer</scene>. ''P. xenovorans'' degrades organic wastes like polychlorinated biphenyls. ''Px''SDR shares less than 37% sequence identity with any known structure and assembles as a prototypical SDR tetramer. As expected, there is some conformational flexibility and differences between the substrate-binding cavity, which explains substrate specificity. Uniquely, the cofactor binding cavity of ''Px''SDR is not well conserved and differs from other SDRs. ''Px''SDR has an additional seven-amino acids that form an additional unique loop within the co-factor binding cavity. Further studies are required to determine how these differences affect the enzymatic functions of the SDR. This project is an educational collaboration between the Seattle Structural Genomics Center for Infectious Disease (SSGCID) and Hampton University where undergraduate students are engaged in structure-function analysis and publication of structures solved by the SSGCID. | + | The high-resolution structure of a putative short-chain reductase from the commercially important bacteria ''Paraburkholderia xenovorans'' is reported (''Px''SDR). The reported apo structure of ''Px''SDR was determined in the monoclinic space group ''P''2<sub>1</sub> as a <scene name='89/899477/Cv/2'>prototypical SDR tetramer</scene>. ''P. xenovorans'' degrades organic wastes like polychlorinated biphenyls. ''Px''SDR shares less than 37% sequence identity with any known structure and assembles as a prototypical SDR tetramer. |
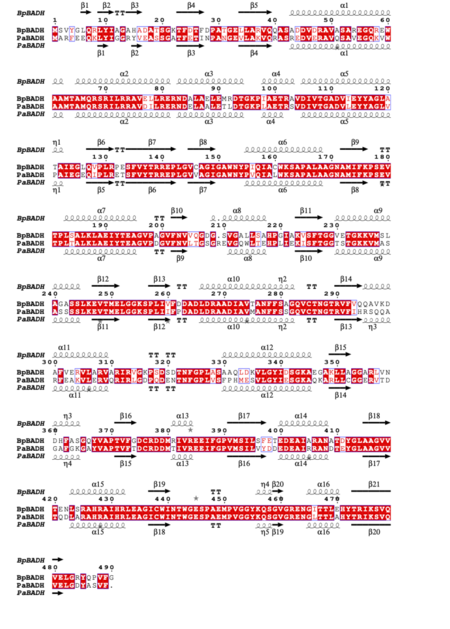
| + | [[Image:Fig2test (1).png|left|450px|thumb|Figure 2 Structural and primary sequence alignment of BpBADH and PaBADH. The secondary structure elements shown are alpha helices (α), 310-helices (η), beta strands (β), and beta turns (TT). Identical residues are shown in white on a red background and conserved residues in red.]] | ||
| + | {{Clear}} | ||
| + | As expected, there is some conformational flexibility and differences between the substrate-binding cavity, which explains substrate specificity. Uniquely, the cofactor binding cavity of ''Px''SDR is not well conserved and differs from other SDRs. ''Px''SDR has an additional seven-amino acids that form an additional unique loop within the co-factor binding cavity. Further studies are required to determine how these differences affect the enzymatic functions of the SDR. This project is an educational collaboration between the Seattle Structural Genomics Center for Infectious Disease (SSGCID) and Hampton University where undergraduate students are engaged in structure-function analysis and publication of structures solved by the SSGCID. | ||
<b>References</b><br> | <b>References</b><br> | ||
Revision as of 13:32, 21 December 2021
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.