We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1644
From Proteopedia
(Difference between revisions)
| Line 80: | Line 80: | ||
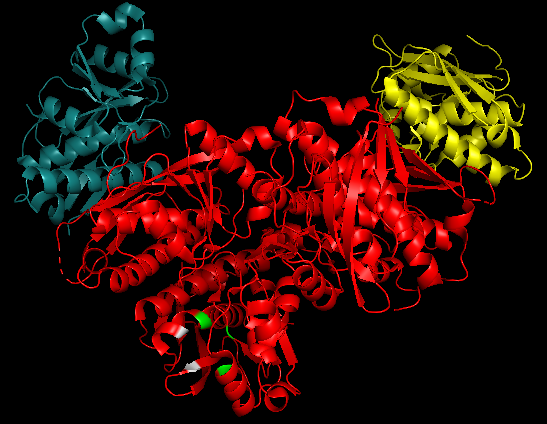
<p align="justify">The '''active site''' represent by 2x36 is composed of''' six [https://en.wikipedia.org/wiki/Protomer protomers]''' in the asymmetric unite. One protomer counts nine [https://en.wikipedia.org/wiki/Beta_sheet b-strands] and seven [https://en.wikipedia.org/wiki/Alpha_helix a-helices]. An analysis of the complex’ structure suggested that '''two pair of protomers''' form A:B and C:D dimers and that the '''two''' remaining ones remain '''uncoupled'''. The dimer interface A:B/C:D is mostly linked by one another through '''hydrophilic interactions''', where the a1-helix is packed against the b3-strand and the loop between b7 and b8 makes inter-subunit contacts with b2<ref>PMID: 20222013</ref>. | <p align="justify">The '''active site''' represent by 2x36 is composed of''' six [https://en.wikipedia.org/wiki/Protomer protomers]''' in the asymmetric unite. One protomer counts nine [https://en.wikipedia.org/wiki/Beta_sheet b-strands] and seven [https://en.wikipedia.org/wiki/Alpha_helix a-helices]. An analysis of the complex’ structure suggested that '''two pair of protomers''' form A:B and C:D dimers and that the '''two''' remaining ones remain '''uncoupled'''. The dimer interface A:B/C:D is mostly linked by one another through '''hydrophilic interactions''', where the a1-helix is packed against the b3-strand and the loop between b7 and b8 makes inter-subunit contacts with b2<ref>PMID: 20222013</ref>. | ||
As all LonA proteins, ''h''Lon catalytic activity relies on a '''Ser-Lys dyad'''. Ser855 on a2 conducts the catalytic cleavage with the assistance of Lys898 on a3 through their [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen-bonding]. The lysine works as a general [https://en.wikipedia.org/wiki/Base_(chemistry) base] along with Thr880 which, in their deprotonated form, abstract the proton from the [https://en.wikipedia.org/wiki/Nucleophile nucleophilic] serine. Those three residues constitute the <scene name='86/868177/Hlonp_catalytic_core/1'>catalytic core</scene>. A characteristic of ''h''LonP is that the [https://en.wikipedia.org/wiki/310_helix 3(10)] helix at the N-terminal end of a2 is able to bring an '''additional residue into the active site''', Asp852. This most likely enables Lys898 [https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa] lowering by creating a [https://en.wikipedia.org/wiki/Hydrophobe hydrophobic] environment, and thus, prevents the dyad to cut off protein substrates. This catalytic '''inactive form''' is also supported by the Asp852 and Trp770 residues that contribute to the <scene name='86/868177/Hlonp_closed_catalitic_core/1'>catalytic site obstruction</scene>. Asp852 removal from the active site through conformational changes enables ''h''Lon to reach an open state that can hydrolyze protein substrate through ATP consumption<ref>PMID: 20222013</ref>.</p> | As all LonA proteins, ''h''Lon catalytic activity relies on a '''Ser-Lys dyad'''. Ser855 on a2 conducts the catalytic cleavage with the assistance of Lys898 on a3 through their [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen-bonding]. The lysine works as a general [https://en.wikipedia.org/wiki/Base_(chemistry) base] along with Thr880 which, in their deprotonated form, abstract the proton from the [https://en.wikipedia.org/wiki/Nucleophile nucleophilic] serine. Those three residues constitute the <scene name='86/868177/Hlonp_catalytic_core/1'>catalytic core</scene>. A characteristic of ''h''LonP is that the [https://en.wikipedia.org/wiki/310_helix 3(10)] helix at the N-terminal end of a2 is able to bring an '''additional residue into the active site''', Asp852. This most likely enables Lys898 [https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa] lowering by creating a [https://en.wikipedia.org/wiki/Hydrophobe hydrophobic] environment, and thus, prevents the dyad to cut off protein substrates. This catalytic '''inactive form''' is also supported by the Asp852 and Trp770 residues that contribute to the <scene name='86/868177/Hlonp_closed_catalitic_core/1'>catalytic site obstruction</scene>. Asp852 removal from the active site through conformational changes enables ''h''Lon to reach an open state that can hydrolyze protein substrate through ATP consumption<ref>PMID: 20222013</ref>.</p> | ||
| - | <br> The figure below shows a 3D simulation of the | + | <br> The figure below shows a 3D simulation of the 2x36 protein, in which the catalytic core (in green) and the catalytic site obstruction (in grey) are highlighted and can be more easily observed. |
[[Image:2x36as.png]] | [[Image:2x36as.png]] | ||
Revision as of 17:40, 20 January 2022
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
2x36 - Structure of the proteolytic domain of the
| |||||||||||
References
- ↑ Matsushima, Y., Takahashi, K., Yue, S., Fujiyoshi, Y., Yoshioka, H., Aihara, M., ... & Kang, D. (2021). Mitochondrial Lon protease is a gatekeeper for proteins newly imported into the matrix. Communications biology, 4(1), 1-13.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Lu, Bin. “Mitochondrial Lon Protease and Cancer.” Advances in Experimental Medicine and Biology 1038 (2017): 173–82. https://doi.org/10.1007/978-981-10-6674-0_12.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ García-Nafría, Javier, Gabriela Ondrovičová, Elena Blagova, Vladimir M Levdikov, Jacob A Bauer, Carolyn K Suzuki, Eva Kutejová, Anthony J Wilkinson, and Keith S Wilson. “Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.” Protein Science : A Publication of the Protein Society 19, no. 5 (May 2010): 987–99. https://doi.org/10.1002/pro.376.
- ↑ Lu, Bin. “Mitochondrial Lon Protease and Cancer.” Advances in Experimental Medicine and Biology 1038 (2017): 173–82. https://doi.org/10.1007/978-981-10-6674-0_12.
- ↑ « The N-terminal domain plays a crucial role in the structure of a full-length human mitochondrial Lon protease | Scientific Reports ». Consulté le 13 janvier 2021. https://www.nature.com/articles/srep33631.
- ↑ He, Lihong, Dongyang Luo, Fan Yang, Chunhao Li, Xuegong Zhang, Haiteng Deng, et Jing-Ren Zhang. « Multiple domains of bacterial and human Lon proteases define substrate selectivity ». Emerging Microbes & Infections 7 (17 août 2018). https://doi.org/10.1038/s41426-018-0148-4.
- ↑ Pomatto, L. C., Carney, C., Shen, B., Wong, S., Halaszynski, K., Salomon, M. P., ... & Tower, J. (2017). The mitochondrial Lon protease is required for age-specific and sex-specific adaptation to oxidative stress. Current Biology, 27(1), 1-15.
- ↑ Kutejová, Eva. « Mitochondrial Lon protease-unique structure and essential function in mammalian cells ». Integrative Cancer Science and Therapeutics 5, nᵒ 6 (2018). https://doi.org/10.15761/ICST.1000296.
- ↑ Voos, Wolfgang, et Karen Pollecker. « The Mitochondrial Lon Protease: Novel Functions off the Beaten Track? » Biomolecules 10, nᵒ 2 (7 février 2020). https://doi.org/10.3390/biom10020253.
- ↑ Coscia, F., & Löwe, J. (2021). Cryo‐EM structure of the full‐length Lon protease from Thermus thermophilus. FEBS letters, 595(21), 2691-2700.
- ↑ He, Lihong, Dongyang Luo, Fan Yang, Chunhao Li, Xuegong Zhang, Haiteng Deng, et Jing-Ren Zhang. « Multiple domains of bacterial and human Lon proteases define substrate selectivity ». Emerging Microbes & Infections 7 (17 août 2018). https://doi.org/10.1038/s41426-018-0148-4.
- ↑ Lu, Bin, Swati Yadav, Parul G. Shah, Tong Liu, Bin Tian, Sebastian Pukszta, Nerissa Villaluna, et al. « Roles for the Human ATP-Dependent Lon Protease in Mitochondrial DNA Maintenance ». Journal of Biological Chemistry 282, nᵒ 24 (15 juin 2007): 17363‑74. https://doi.org/10.1074/jbc.M611540200.
- ↑ Kereiche S, Kovacik L, Bednar J, Pevala V, Kunova N, Ondrovicova G, Bauer J, Ambro L, Bellova J, Kutejova E, Raska I. The N-terminal domain plays a crucial role in the structure of a full-length human mitochondrial Lon protease. Sci Rep. 2016 Sep 16;6:33631. doi: 10.1038/srep33631. PMID:27632940 doi:http://dx.doi.org/10.1038/srep33631
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Wang, N, S Gottesman, M C Willingham, M M Gottesman, and M R Maurizi. “A Human Mitochondrial ATP-Dependent Protease That Is Highly Homologous to Bacterial Lon Protease.” Proceedings of the National Academy of Sciences 90, no. 23 (December 1, 1993): 11247–51. https://doi.org/10.1073/pnas.90.23.11247.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mhttps://proteopedia.org/wiki/skins/common/images/button_extlink.pngitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Lon Protease Preferentially Degrades Oxidized Mitochondrial Aconitase by an ATP-Stimulated Mechanism.” Nature Cell Biology 4, no. 9 (September 2002): 674–80. https://doi.org/10.1038/ncb836.