Sandbox reserved 1651
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='6Z1N' size='360' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='6Z1N' size='360' side='right' caption='Caption for this structure' scene=''> | ||
| - | Cis-Prenyltransferases (cis-PTs) is a large enzyme family which is well conserved in all areas of life. Cis-PTs catalyze condensation reactions of [https://en.wikipedia.org/wiki/Isopentenyl_pyrophosphate isopentenyl pyrophosphate (IPP)] and produce linear polyprenyl diphosphate. The length of this [https://en.wikipedia.org/wiki/Terpenoid isoprenoid] carbon chain ranges from short molecules like [https://en.wikipedia.org/wiki/Geranyl_pyrophosphate geranyl diphosphate] (C10) to natural rubber (C>10’000). The human cis-Prenyltransferase Complex (hcis-PT) plays an essential role in protein [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]. It synthesizes the precursor of [https://en.wikipedia.org/wiki/Glycosyl glycosyl] carrier [https://en.wikipedia.org/wiki/Dolichol dolichol]-phosphate. Mutations in genes coding for hcis-PT can cause serious diseases, such as [https://en.wikipedia.org/wiki/Retinitis_pigmentosa retinitis pigmentosa].[1] | + | <p align="justify">Cis-Prenyltransferases (cis-PTs) is a large enzyme family which is well conserved in all areas of life. Cis-PTs catalyze condensation reactions of [https://en.wikipedia.org/wiki/Isopentenyl_pyrophosphate isopentenyl pyrophosphate (IPP)] and produce linear polyprenyl diphosphate. The length of this [https://en.wikipedia.org/wiki/Terpenoid isoprenoid] carbon chain ranges from short molecules like [https://en.wikipedia.org/wiki/Geranyl_pyrophosphate geranyl diphosphate] (C10) to natural rubber (C>10’000). The human cis-Prenyltransferase Complex (hcis-PT) plays an essential role in protein [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]. It synthesizes the precursor of [https://en.wikipedia.org/wiki/Glycosyl glycosyl] carrier [https://en.wikipedia.org/wiki/Dolichol dolichol]-phosphate. Mutations in genes coding for hcis-PT can cause serious diseases, such as [https://en.wikipedia.org/wiki/Retinitis_pigmentosa retinitis pigmentosa].[1] </p> |
== Structure == | == Structure == | ||
| Line 9: | Line 9: | ||
=== The hcis-PT subunits === | === The hcis-PT subunits === | ||
| - | The hcis-PT is a tetramer formed by the assembly of a dimer of heterodimers. The hcis-PT is composed of two catalytically active [https://en.wikipedia.org/wiki/Dehydrodolichyl_diphosphate_synthase dehydrodolichyl diphosphate synthases] (<scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene>) and the Nogo-B receptor (<scene name='87/872232/Ngbr_aliases_nus1/1'>NgBR</scene>). The NgBR can be split into two domains: a C-terminal pseudo [https://en.wikipedia.org/wiki/Prenyltransferase prenyltransferase] domain that interacts with DHDDS and an N-terminal transmembrane domain that is anchored in the endoplasmic reticulum. The NgBR C-terminus encompasses the RxG pattern which might have an important role in hcis-PT activity and which is not present in NUS1 ([[6jcn]]), the yeast homolog of NgBR. The structure of DHDDS can be divided into three domains: a <scene name='87/872232/Dhdds_ctd/1'>C-terminal domain</scene> (residues 251–333), a <scene name='87/872232/Dhdds_n-terminal_domain/1'>N-terminal domain</scene> (residues 1–26) and a <scene name='87/872232/Dhdds_catalytic_domain/1'>canonical catalytic cis-PT homology domain</scene> (residues 27–250). The cis-PT homology domain heterodimerizes with NgBR. The tetramer is formed by heterotypic interactions of the "turn" region with NgBR and by homotypic interactions between DHDDS. The whole complex is headed towards the cytosol. | + | <p align="justify"> The hcis-PT is a tetramer formed by the assembly of a dimer of heterodimers. The hcis-PT is composed of two catalytically active [https://en.wikipedia.org/wiki/Dehydrodolichyl_diphosphate_synthase dehydrodolichyl diphosphate synthases] (<scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene>) and the Nogo-B receptor (<scene name='87/872232/Ngbr_aliases_nus1/1'>NgBR</scene>). The NgBR can be split into two domains: a C-terminal pseudo [https://en.wikipedia.org/wiki/Prenyltransferase prenyltransferase] domain that interacts with DHDDS and an N-terminal transmembrane domain that is anchored in the endoplasmic reticulum. The NgBR C-terminus encompasses the RxG pattern which might have an important role in hcis-PT activity and which is not present in NUS1 ([[6jcn]]), the yeast homolog of NgBR. The structure of DHDDS can be divided into three domains: a <scene name='87/872232/Dhdds_ctd/1'>C-terminal domain</scene> (residues 251–333), a <scene name='87/872232/Dhdds_n-terminal_domain/1'>N-terminal domain</scene> (residues 1–26) and a <scene name='87/872232/Dhdds_catalytic_domain/1'>canonical catalytic cis-PT homology domain</scene> (residues 27–250). The cis-PT homology domain heterodimerizes with NgBR. The tetramer is formed by heterotypic interactions of the "turn" region with NgBR and by homotypic interactions between DHDDS. The whole complex is headed towards the cytosol. </p> |
=== The active site organization === | === The active site organization === | ||
| - | [https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate Farnesyl diphosphate (FPP)] and [https://en.wikipedia.org/wiki/Magnesium#Chemical_properties Mg<sup>2+</sup>] are only detected in the active site of DHDDS and only the active site of DHDDS is required for the catalytic activity. NgBR induces an increase in the expression and activity of the complex, but itself has no catalytic activity. Indeed, NgBR active site does not have a visible substrate-binding cavity. The 3 [https://en.wikipedia.org/wiki/Beta_sheet β-strands] and 2 [https://en.wikipedia.org/wiki/Alpha_helix α-helices] are packaged via [https://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic interactions], so there is no longer a cavity and it is devoid of water. | + | <p align="justify">[https://en.wikipedia.org/wiki/Farnesyl_pyrophosphate Farnesyl diphosphate (FPP)] and [https://en.wikipedia.org/wiki/Magnesium#Chemical_properties Mg<sup>2+</sup>] are only detected in the active site of DHDDS and only the active site of DHDDS is required for the catalytic activity. NgBR induces an increase in the expression and activity of the complex, but itself has no catalytic activity. Indeed, NgBR active site does not have a visible substrate-binding cavity. The 3 [https://en.wikipedia.org/wiki/Beta_sheet β-strands] and 2 [https://en.wikipedia.org/wiki/Alpha_helix α-helices] are packaged via [https://en.wikipedia.org/wiki/Hydrophobic_effect hydrophobic interactions], so there is no longer a cavity and it is devoid of water. </p> |
| Line 19: | Line 19: | ||
== Catalytical activity of the human cis-prenyltransferases == | == Catalytical activity of the human cis-prenyltransferases == | ||
| - | The catalytical domain of DHDDS is homologous to [[undecaprenyl pyrophosphate synthase]] (UPPS) with 2 α-helices and four β-strands within each monomer. The active site is shaped by a <scene name='87/872232/Superficial_polar_region/1'>superficial polar region</scene> that stabilizes the interaction between IPP and a <scene name='87/872232/Hydrophobic_tunnel/1'>deep hydrophobic tunnel</scene> which accommodate the elongating carbon chain. In the active site, there are two substrate-binding sites, an S1, and an S2 site. The <scene name='87/872232/S1_site/2'>S1 site</scene> binds the initiator substrate FPP. It additionally interacts with Mg<sup>2+</sup> ions which are crucial for IPP hydrolysis during the condensation reaction. The Mg<sup>2+</sup> is stabilized and <scene name='87/872232/Octahedrally_coordinated_mg/3'>octahedrally coordinated</scene> by three surrounding water molecules, two oxygens of the [https://en.wikipedia.org/wiki/Pyrophosphate pyrophosphate] and one [https://en.wikipedia.org/wiki/Carboxylate carboxylate] oxygen of D34, that ensures the biological function of the protein. <scene name='87/872232/S2_site/1'>S2 site</scene> binds the IPP molecule which will be employed for chain elongation. The C-terminus of NgBR (RxG) is directly involved in forming the superficial polar layer and enables the formation of S1 and S2. In fact, at the S1 site, there are two polar interaction networks between NgBR and DHDDS. At <scene name='87/872232/S2_site/1'>S2 site</scene>, the backbone nitrogen atoms directly coordinate the phosphate molecule. | + | <p align="justify"> The catalytical domain of DHDDS is homologous to [[undecaprenyl pyrophosphate synthase]] (UPPS) with 2 α-helices and four β-strands within each monomer. The active site is shaped by a <scene name='87/872232/Superficial_polar_region/1'>superficial polar region</scene> that stabilizes the interaction between IPP and a <scene name='87/872232/Hydrophobic_tunnel/1'>deep hydrophobic tunnel</scene> which accommodate the elongating carbon chain. In the active site, there are two substrate-binding sites, an S1, and an S2 site. The <scene name='87/872232/S1_site/2'>S1 site</scene> binds the initiator substrate FPP. It additionally interacts with Mg<sup>2+</sup> ions which are crucial for IPP hydrolysis during the condensation reaction. The Mg<sup>2+</sup> is stabilized and <scene name='87/872232/Octahedrally_coordinated_mg/3'>octahedrally coordinated</scene> by three surrounding water molecules, two oxygens of the [https://en.wikipedia.org/wiki/Pyrophosphate pyrophosphate] and one [https://en.wikipedia.org/wiki/Carboxylate carboxylate] oxygen of D34, that ensures the biological function of the protein. <scene name='87/872232/S2_site/1'>S2 site</scene> binds the IPP molecule which will be employed for chain elongation. The C-terminus of NgBR (RxG) is directly involved in forming the superficial polar layer and enables the formation of S1 and S2. In fact, at the S1 site, there are two polar interaction networks between NgBR and DHDDS. At <scene name='87/872232/S2_site/1'>S2 site</scene>, the backbone nitrogen atoms directly coordinate the phosphate molecule. </p> |
=== The elongation reaction in the hydrophobic active-site tunnel === | === The elongation reaction in the hydrophobic active-site tunnel === | ||
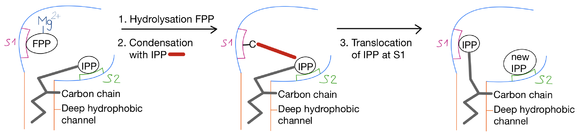
| - | The cis-PT catalyzes the chain elongation of FPP by successive head-to-tail condensation with a specific number of IPP to form linear lipids with designated chain lengths. The reaction is as follows: | + | <p align="justify">The cis-PT catalyzes the chain elongation of FPP by successive head-to-tail condensation with a specific number of IPP to form linear lipids with designated chain lengths. The reaction is as follows: |
| - | IPP headgroups are bound to the enzyme at the superficial polar region while the carbon chains point toward the <scene name='87/872232/Hydrophobic_tunnel/1'>deep hydrophobic tunnel</scene>. First, the pyrophosphate group (FPP) of the initiatory substrate, which interacts with a Mg<sup>2+</sup> ion, is hydrolyzed at the <scene name='87/872232/S1_site/2'>S1 site</scene>. Then, the condensation of the remaining carbon with the IPP from the S2 site follows. The elongated product translocate to the S1, so the carbon chain goes into the hydrophobic tunnel of the active site. In the end, a new IPP molecule binds to the S2 site and the cycle is repeated until the active site can no longer accommodate the long-chain [https://en.wikipedia.org/wiki/Terpenoid isoprenoid]. | + | <br> |
| + | IPP headgroups are bound to the enzyme at the superficial polar region while the carbon chains point toward the <scene name='87/872232/Hydrophobic_tunnel/1'>deep hydrophobic tunnel</scene>. First, the pyrophosphate group (FPP) of the initiatory substrate, which interacts with a Mg<sup>2+</sup> ion, is hydrolyzed at the <scene name='87/872232/S1_site/2'>S1 site</scene>. Then, the condensation of the remaining carbon with the IPP from the S2 site follows. The elongated product translocate to the S1, so the carbon chain goes into the hydrophobic tunnel of the active site. In the end, a new IPP molecule binds to the S2 site and the cycle is repeated until the active site can no longer accommodate the long-chain [https://en.wikipedia.org/wiki/Terpenoid isoprenoid]. </p> | ||
[[Image:Elongation reaction.png | thumb | center | 580px | '''Elongation reaction''' ]] | [[Image:Elongation reaction.png | thumb | center | 580px | '''Elongation reaction''' ]] | ||
| Line 30: | Line 31: | ||
=== Regulation of the product length === | === Regulation of the product length === | ||
| - | The hydrophobic tunnel of DHDDS is formed by 2 α-helix and 4 β-strands. The opening between the 2 α-helices is larger in DHDDS compared to short and medium-chain cis-PT. Larger is the diameter, better is the accommodation of a longer product. | + | <p align="justify"> The hydrophobic tunnel of DHDDS is formed by 2 α-helix and 4 β-strands. The opening between the 2 α-helices is larger in DHDDS compared to short and medium-chain cis-PT. Larger is the diameter, better is the accommodation of a longer product. </p> |
| Line 39: | Line 40: | ||
=== Product of the N-glycosylation === | === Product of the N-glycosylation === | ||
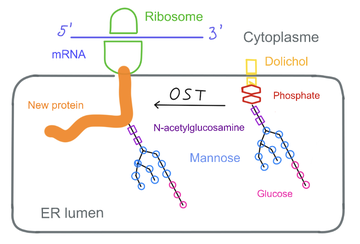
| - | The [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]is a post-translational modification realized in the endoplasmic reticulum of the cell. This process consists in linking a [https://en.wikipedia.org/wiki/Glycan glycan] to a protein, which provide the biological protein fonction and [https://en.wikipedia.org/wiki/Protein_folding folding]. The hcis-PT produces DHDD (dehydrodolichyl diphosphate), an important precursor molecule for the dolichol-phosphate lipid carrier needed in the [https://fr.wikipedia.org/wiki/N-glycosylation N-glycosylation] reaction. | + | <p align="justify"> The [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]is a post-translational modification realized in the endoplasmic reticulum of the cell. This process consists in linking a [https://en.wikipedia.org/wiki/Glycan glycan] to a protein, which provide the biological protein fonction and [https://en.wikipedia.org/wiki/Protein_folding folding]. The hcis-PT produces DHDD (dehydrodolichyl diphosphate), an important precursor molecule for the dolichol-phosphate lipid carrier needed in the [https://fr.wikipedia.org/wiki/N-glycosylation N-glycosylation] reaction. [2]</p> |
| - | [[image:N-glycosylation.png | thumb | center|360px| upright=10|'''N-glycosylation'''] | + | [[image:N-glycosylation.png | thumb | center|360px| upright=10|'''N-glycosylation''']] |
=== Disease comprehension === | === Disease comprehension === | ||
| - | The [https://en.wikipedia.org/wiki/Retinitis_pigmentosa Retinitis pigmentosa] is a [https://en.wikipedia.org/wiki/Genetic_disorder hereditary disease]. Patients lose progressively parts of their sight: night vision as a teenager, side vision as a young adult, and central vision in later life. This is due to the progressive decrease of [https://en.wikipedia.org/wiki/Rod_cell rod] and [https://en.wikipedia.org/wiki/Cone_cell cone] [https://en.wikipedia.org/wiki/Photoreceptor_cell photoreceptor cells] which provide colored vision. Scientists observed that symptoms appear many years after the beginning of the photoreceptor’s [https://en.wikipedia.org/wiki/Degenerative_disease degeneration] in most cases. Retinitis pigmentosa can be encoded by many genes. More than 45 have been identified but these genes concern only 60% of sick patients. Therefore 40% of cases of Retinitis pigmentosa are unidentified genes. Until today there is no cure. However, studies showed a slow down of the disease with the intake of [https://en.wikipedia.org/wiki/Vitamin_A vitamin A], [https://en.wikipedia.org/wiki/Palmitic_acid palmitate] foods and [https://en.wikipedia.org/wiki/Omega-3_fatty_acid omega-3]-rich fish [3]. | + | <p align="justify">The [https://en.wikipedia.org/wiki/Retinitis_pigmentosa Retinitis pigmentosa] is a [https://en.wikipedia.org/wiki/Genetic_disorder hereditary disease]. Patients lose progressively parts of their sight: night vision as a teenager, side vision as a young adult, and central vision in later life. This is due to the progressive decrease of [https://en.wikipedia.org/wiki/Rod_cell rod] and [https://en.wikipedia.org/wiki/Cone_cell cone] [https://en.wikipedia.org/wiki/Photoreceptor_cell photoreceptor cells] which provide colored vision. Scientists observed that symptoms appear many years after the beginning of the photoreceptor’s [https://en.wikipedia.org/wiki/Degenerative_disease degeneration] in most cases. Retinitis pigmentosa can be encoded by many genes. More than 45 have been identified but these genes concern only 60% of sick patients. Therefore 40% of cases of Retinitis pigmentosa are unidentified genes. Until today there is no cure. However, studies showed a slow down of the disease with the intake of [https://en.wikipedia.org/wiki/Vitamin_A vitamin A], [https://en.wikipedia.org/wiki/Palmitic_acid palmitate] foods and [https://en.wikipedia.org/wiki/Omega-3_fatty_acid omega-3]-rich fish [3]. </p> |
| - | For instance, <scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene> missense mutations can provoke an [https://en.wikipedia.org/w/index.php?title=Autosomal_Recessive&redirect=no autosomal recessive] Retinitis pigmentosa (arRP). These mutations concern the S1 and S2 sites of the active site where pyrophosphate can bind and most of the mutations related to the disease impact directly the substrate binding, according to scientists. For one mutation, <scene name='87/872232/K42/3'>K42E</scene>, it is more complicated. Scientists remarked that, in the <scene name='87/872232/Salt_bridge_k42-e234/1'>wild type, R38 points toward the active-site cavity, while K42 and E234 form a stable salt bridge</scene> (short distance). The mutation K42E provokes in the protein scale, hypothetically, an interaction with the adjacent active-site residues R38 positively charged. So R38 points away from the active site cavity, forming a new stable [[salt bridge]] with the mutant E42. The distance between K42E and E234 is longer due to the charge repulsion. Finally, experiments proved that aberrant polar networks are due to the K42E mutation, disturbing the active-site residues which can’t interact with the substrate and leading to a decrease of the catalytic activity [1]. | + | <p align="justify"> For instance, <scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene> missense mutations can provoke an [https://en.wikipedia.org/w/index.php?title=Autosomal_Recessive&redirect=no autosomal recessive] Retinitis pigmentosa (arRP). These mutations concern the S1 and S2 sites of the active site where pyrophosphate can bind and most of the mutations related to the disease impact directly the substrate binding, according to scientists. For one mutation, <scene name='87/872232/K42/3'>K42E</scene>, it is more complicated. Scientists remarked that, in the <scene name='87/872232/Salt_bridge_k42-e234/1'>wild type, R38 points toward the active-site cavity, while K42 and E234 form a stable salt bridge</scene> (short distance). The mutation K42E provokes in the protein scale, hypothetically, an interaction with the adjacent active-site residues R38 positively charged. So R38 points away from the active site cavity, forming a new stable [[salt bridge]] with the mutant E42. The distance between K42E and E234 is longer due to the charge repulsion. Finally, experiments proved that aberrant polar networks are due to the K42E mutation, disturbing the active-site residues which can’t interact with the substrate and leading to a decrease of the catalytic activity [1]. </p> |
Current revision
Heterotetrameric Cis-Prenyltransferase Complex
| |||||||||||