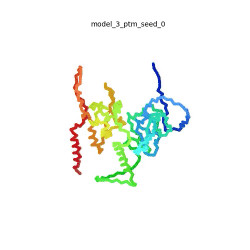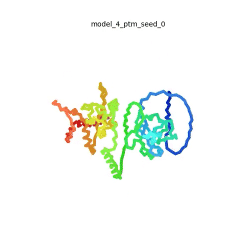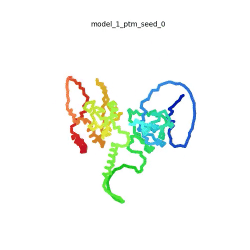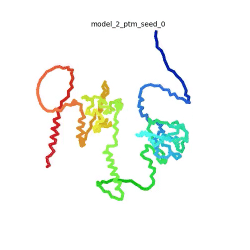
SARS-CoV-2 protein N
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | ==SARS CoV-2 Protein N== | |
| + | <StructureSection load='' size='350' side='right' scene='90/902745/Sars-cov-2_protein_n/3' caption='SARS-CoV-2 Protein N'> | ||
{{Theoretical_model}} | {{Theoretical_model}} | ||
| + | {{Template:ModelConfidence}} | ||
| + | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein N (UniProt ID: QHD43423) color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 5<ref name="MIT_ColabFold"> https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb </ref>. | ||
== Function == | == Function == | ||
The primary function of the Nucleocapsid protein (N-protein) is to package the viral genome into a helical ribonucleoprotein (RNP) complex that protects the genomic RNA, and bind it tightly during the virus’s journey to a new host. <ref name="Chang"> PMID:24418573 </ref>N-proteins are necessary for viral RNA transcription and replication and are also regulating these processes. <ref name="Kang"> https://www.sciencedirect.com/science/article/pii/S2211383520305505 </ref>The packaging of the RNA into the RNP complex is a fundamental part of the RNA assembly in infected cells <ref name="Kang"/>. N-proteins are also involved in the replication cycle and have influence on the hosts cellular response to viral infection. <ref name="Chang"/> | The primary function of the Nucleocapsid protein (N-protein) is to package the viral genome into a helical ribonucleoprotein (RNP) complex that protects the genomic RNA, and bind it tightly during the virus’s journey to a new host. <ref name="Chang"> PMID:24418573 </ref>N-proteins are necessary for viral RNA transcription and replication and are also regulating these processes. <ref name="Kang"> https://www.sciencedirect.com/science/article/pii/S2211383520305505 </ref>The packaging of the RNA into the RNP complex is a fundamental part of the RNA assembly in infected cells <ref name="Kang"/>. N-proteins are also involved in the replication cycle and have influence on the hosts cellular response to viral infection. <ref name="Chang"/> | ||
== Structure description == | == Structure description == | ||
| - | The N-protein consists of 419 amino acids and can be divided into two folded domains and three disordered regions. The three disordered regions dynamically change their conformation and are very flexible, which allows them to rotate and enable the binding to macromolecules like RNA. The two folded domains on the other hand are well structured and have thus been modelled using X-Ray diffraction and NMR. <ref name="Cubuk"> https://www.biorxiv.org/content/10.1101/2020.06.17.158121v1.full </ref> | + | [[Image:SARSCoV2_protein_N_250.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein N''', ''rainbow'' color coded N-C<ref name="MIT_ColabFold"/>.]]<br />The N-protein consists of 419 amino acids and can be divided into two folded domains and three disordered regions. The three disordered regions dynamically change their conformation and are very flexible, which allows them to rotate and enable the binding to macromolecules like RNA. The two folded domains on the other hand are well structured and have thus been modelled using X-Ray diffraction and NMR. <ref name="Cubuk"> https://www.biorxiv.org/content/10.1101/2020.06.17.158121v1.full </ref> |
=== N-terminal flexible arm === | === N-terminal flexible arm === | ||
| Line 40: | Line 43: | ||
[[Coronavirus_Disease 2019 (COVID-19)]] | [[Coronavirus_Disease 2019 (COVID-19)]] | ||
__NOTOC__ | __NOTOC__ | ||
| - | </SX> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | </StructureSection> | ||
Revision as of 12:12, 3 February 2022
SARS CoV-2 Protein N
| |||||||||||