SARS-CoV-2 protein NSP11
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene=' | + | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene='90/903539/Alphafold2_nsp11/1'> |
{{Theoretical_model}} {{Template:ModelConfidence}} | {{Theoretical_model}} {{Template:ModelConfidence}} | ||
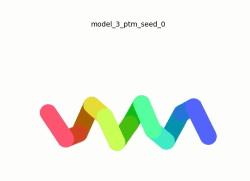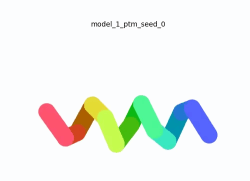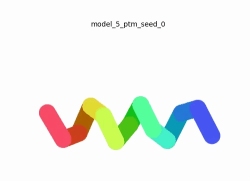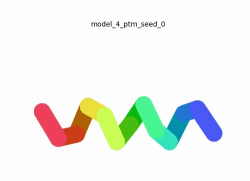
To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, NCBI ID [https://www.ncbi.nlm.nih.gov/protein/YP_009725302.1 YP_009725302]) color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 3<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein NSP11 (length = 13 amino acids, NCBI ID [https://www.ncbi.nlm.nih.gov/protein/YP_009725302.1 YP_009725302]) color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 3<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. | ||
Revision as of 14:49, 8 February 2022
| |||||||||||
References
- ↑ 1.0 1.1 1.2 MIT ColabFold
- ↑ Modeling of the SARS-COV-2 Genome
- ↑ Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res. 2020 Apr 3;19(4):1351-1360. doi: 10.1021/acs.jproteome.0c00129., Epub 2020 Mar 24. PMID:32200634 doi:http://dx.doi.org/10.1021/acs.jproteome.0c00129