3g0h
From Proteopedia
| Line 1: | Line 1: | ||
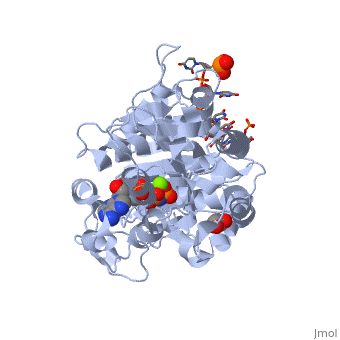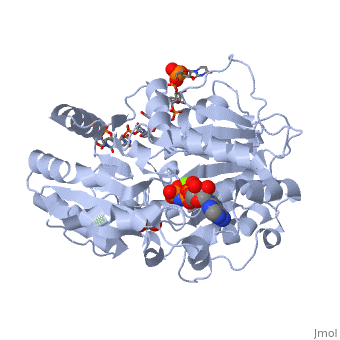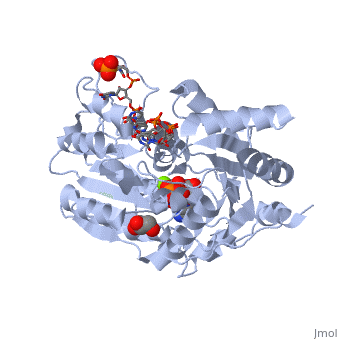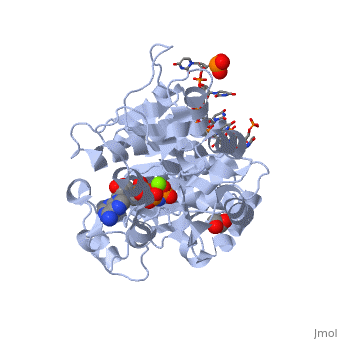
==Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA== | ==Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA== | ||
| - | <StructureSection load='3g0h' size='340' side='right' caption='[[3g0h]], [[Resolution|resolution]] 2.70Å' scene=''> | + | <StructureSection load='3g0h' size='340' side='right'caption='[[3g0h]], [[Resolution|resolution]] 2.70Å' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[3g0h]] is a 2 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[3g0h]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Human Human]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3G0H OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3G0H FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat"><scene name='pdbligand=ANP:PHOSPHOAMINOPHOSPHONIC+ACID-ADENYLATE+ESTER'>ANP</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene></td></tr> | + | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ANP:PHOSPHOAMINOPHOSPHONIC+ACID-ADENYLATE+ESTER'>ANP</scene>, <scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene></td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[3ews|3ews]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[3ews|3ews]]</div></td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">DBP5, DDX19, DDX19B ([ | + | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">DBP5, DDX19, DDX19B ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 HUMAN])</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3g0h FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3g0h OCA], [https://pdbe.org/3g0h PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3g0h RCSB], [https://www.ebi.ac.uk/pdbsum/3g0h PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3g0h ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/DD19B_HUMAN DD19B_HUMAN]] ATP-dependent RNA helicase involved in mRNA export from the nucleus. Rather than unwinding RNA duplexes, DDX19B functions as a remodeler of ribonucleoprotein particles, whereby proteins bound to nuclear mRNA are dissociated and replaced by cytoplasmic mRNA binding proteins. |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 32: | Line 32: | ||
==See Also== | ==See Also== | ||
| - | + | *[[Helicase 3D structures|Helicase 3D structures]] | |
| - | + | ||
| - | *[[Helicase|Helicase]] | + | |
== References == | == References == | ||
<references/> | <references/> | ||
| Line 40: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Human]] | [[Category: Human]] | ||
| + | [[Category: Large Structures]] | ||
[[Category: Arrowsmith, C H]] | [[Category: Arrowsmith, C H]] | ||
[[Category: Berg, S Van Den]] | [[Category: Berg, S Van Den]] | ||
Revision as of 07:52, 9 March 2022
Human dead-box RNA helicase DDX19, in complex with an ATP-analogue and RNA
| |||||||||||
Categories: Human | Large Structures | Arrowsmith, C H | Berg, S Van Den | Berglund, H | Bountra, C | Collins, R | Dahlgren, L G | Edwards, A M | Flodin, S | Flores, A | Graslund, S | Hammarstrom, M | Johansson, A | Johansson, I | Karlberg, T | Kotenyova, T | Lehtio, L | Moche, M | Nilsson, M E | Nordlund, P | Nyman, T | Persson, C | Structural genomic | Sagemark, J | Schuler, H | Schutz, P | Siponen, M I | Thorsell, A G | Tresaugues, L | Weigelt, J | Welin, M | Wisniewska, M | Atp-binding | Dbp5 | Helicase | Hydrolase | Hydrolase-rna complex | Membrane | Mrna transport | Nuclear pore complex | Nucleotide-binding | Nucleus | Phosphoprotein | Polyuracil | Protein transport | Protein-rna complex | Rna-binding | Sgc | Translocation | Transport