This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
<scene name='90/904325/Ras_nf1_complex/2'>NF interacts with Ras to form a complex. R68 assists N61 in catalysis.</scene> | <scene name='90/904325/Ras_nf1_complex/2'>NF interacts with Ras to form a complex. R68 assists N61 in catalysis.</scene> | ||
===Mechanism of Ras Coupled with Neurofibromin=== | ===Mechanism of Ras Coupled with Neurofibromin=== | ||
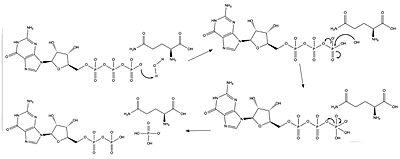
| - | The RasGAP interactions that occur when neurofibromin's GAP domain and Ras are bound have two critical catalytic components. The first is the arginine finger | + | The RasGAP interactions that occur when neurofibromin's GAP domain and Ras are bound have two critical catalytic components. The first is the <scene name='90/904325/Arginine_finger_and_gtp/1'>the arginine finger</scene> of the NF protein, which stabilizes the catalytic glutamine, as well as stabilizing the transition state of the phosphoryl transfer<ref name= ''Scheffzek''>PMID:30104198</ref>. The second component is the catalytic glutamine of the Ras protein, which stabilizes the nucleophilic water as it attacks the third phosphate group in the GTP molecule as shown in Figure 1. |
| - | [[Image:Ras mechanism.jpg|400 px|right|thumb|Figure | + | [[Image:Ras mechanism.jpg|400 px|right|thumb|Figure 1: The catalytic glutamine stabilizes the nucleophilic water.]] |
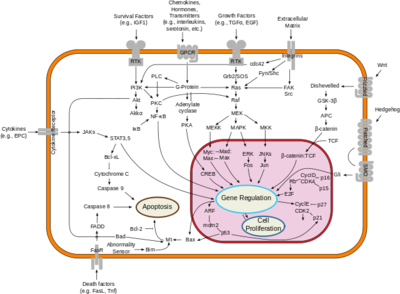
===Downstream Effects=== | ===Downstream Effects=== | ||
[[Image:Signal_transduction_pathways.png|400 px|right|thumb|Figure X; By cybertory - This file was derived from: Signal transduction v1.png, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=12081090]] | [[Image:Signal_transduction_pathways.png|400 px|right|thumb|Figure X; By cybertory - This file was derived from: Signal transduction v1.png, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=12081090]] | ||
Revision as of 20:12, 26 March 2022
| |||||||||||
References
- ↑ Scheffzek K, Shivalingaiah G. Ras-Specific GTPase-Activating Proteins-Structures, Mechanisms, and Interactions. Cold Spring Harb Perspect Med. 2019 Mar 1;9(3). pii: cshperspect.a031500. doi:, 10.1101/cshperspect.a031500. PMID:30104198 doi:http://dx.doi.org/10.1101/cshperspect.a031500
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky