Sandbox Reserved 1716
From Proteopedia
(Difference between revisions)
| Line 38: | Line 38: | ||
==Significant Cysteines== | ==Significant Cysteines== | ||
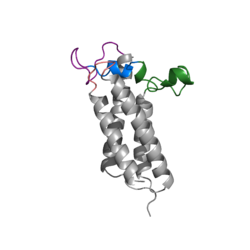
| - | A set of four cysteines is consistently conserved in all VKOR homologs. In the human homolog (HsVKOR) these cysteines are Cys43, Cys51, Cys132, and Cys 135. In the Pufferfish homolog (TrVKORL) these cysteines, due to Cryo-EM differences,are Cys52, Cys55, Cys141, and Cys144. These cysteines are the key factor that allow for Vitamin K Epoxide Reductase to perform its function, which is to open the epoxide ring on Vitamin K Epoxide in order to re-make Vitamin K Quinone. In the closed conformation, that is induced when Vitamin K binds in the hydrophobic pocket, Cys-132 binds to Cys-51 and Cys-135 will bind to the 3' hydroxyl group on Vitamin K Epoxide, which allows for the electron transfer to open up the epoxide ring. | + | A set of four cysteines is consistently conserved in all VKOR homologs. In the human homolog (HsVKOR) these cysteines are Cys43, Cys51, Cys132, and Cys 135. <scene name='90/904321/Cysteines/3'>Significant Cysteines</scene> In the Pufferfish homolog (TrVKORL) these cysteines, due to Cryo-EM differences,are Cys52, Cys55, Cys141, and Cys144. These cysteines are the key factor that allow for Vitamin K Epoxide Reductase to perform its function, which is to open the epoxide ring on Vitamin K Epoxide in order to re-make Vitamin K Quinone. In the closed conformation, that is induced when Vitamin K binds in the hydrophobic pocket, Cys-132 binds to Cys-51 and Cys-135 will bind to the 3' hydroxyl group on Vitamin K Epoxide, which allows for the electron transfer to open up the epoxide ring. |
<scene name='90/904321/Cys52disulfidecys55/3'>Disulfide Bond Cysteine52-Cysteine55 TrVKORL</scene> | <scene name='90/904321/Cys52disulfidecys55/3'>Disulfide Bond Cysteine52-Cysteine55 TrVKORL</scene> | ||
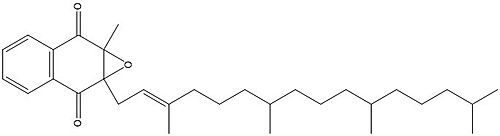
==Vitamin K Epoxide== | ==Vitamin K Epoxide== | ||
Revision as of 14:43, 28 March 2022
Vitamin K Epoxide Reductase
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677