We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1706
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
[https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin. Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MARP/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Mutations that cause a decrease in activity of neurofibromin cause tumors to grow along your nerves. As neurofibromin is ubiquitously expressed throughout the body, these tumors can grow anywhere. Neurofibromin is located in the [https://en.wikipedia.org/wiki/Cytosol cytosol] of the cell but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to bind to [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was detemed through high-resolution single particles [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy] to understand the overall structure and the different domains. [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions. X-Ray crystallography] experiments found inconsistency in the structural determination.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | [https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin. Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MARP/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Mutations that cause a decrease in activity of neurofibromin cause tumors to grow along your nerves. As neurofibromin is ubiquitously expressed throughout the body, these tumors can grow anywhere. Neurofibromin is located in the [https://en.wikipedia.org/wiki/Cytosol cytosol] of the cell but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to bind to [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was detemed through high-resolution single particles [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy] to understand the overall structure and the different domains. [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions. X-Ray crystallography] experiments found inconsistency in the structural determination.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
==Function== | ==Function== | ||
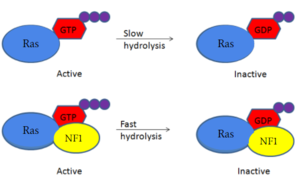
| - | Neurofibromin is a [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] that binds to [https://en.wikipedia.org/wiki/Ras_GTPase Ras], a [https://en.wikipedia.org/wiki/GTPase GTPase], to increase the hydrolysis of GTP to GDP. This inactivates the cell signaling of Ras until another [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] can replace the [https://en.wikipedia.org/wiki/Guanosine_diphosphate GDP] from the cytosol. Neurofibromin and Ras binding is possible in only the <scene name='90/904311/Open_conformation/2'>open conformation</scene> of Neurofibromin. The mechanism is shown in figure 1 and displays the slow hydrolysis of GTP bound to Ras and the fast hydrolysis of GTP when bound to Neurofibromin. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | Neurofibromin is a [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] that binds to [https://en.wikipedia.org/wiki/Ras_GTPase Ras], a [https://en.wikipedia.org/wiki/GTPase GTPase], to increase the hydrolysis of GTP to GDP. This inactivates the cell signaling of Ras until another [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] can replace the [https://en.wikipedia.org/wiki/Guanosine_diphosphate GDP] from the cytosol. Neurofibromin and Ras binding is possible in only the <scene name='90/904311/Open_conformation/2'>open conformation</scene> of Neurofibromin. The mechanism is shown in figure 1 and displays the slow hydrolysis of GTP bound to Ras and the fast hydrolysis of GTP when bound to Neurofibromin. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> [[Image:RasNeurofibrominMech.PNG|300px|left|thumb|blah]] |
==Structure== | ==Structure== | ||
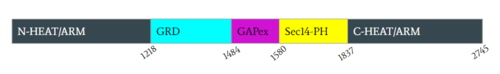
Neurofibromin is a [https://en.wikipedia.org/wiki/Protein_dimer protein dimer] that exists in the<scene name='90/904311/Closed_conformation/3'>closed</scene> and <scene name='90/904311/Open_conformation/2'>open</scene> conformation. Each [https://en.wikipedia.org/wiki/Protomer protomer] contains a GRD, Sec14-PH, and a GAPex domain located on a HEAT N-C arm. Ras binds to the GRD site with Arg1276 being the critical residue for binding. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | Neurofibromin is a [https://en.wikipedia.org/wiki/Protein_dimer protein dimer] that exists in the<scene name='90/904311/Closed_conformation/3'>closed</scene> and <scene name='90/904311/Open_conformation/2'>open</scene> conformation. Each [https://en.wikipedia.org/wiki/Protomer protomer] contains a GRD, Sec14-PH, and a GAPex domain located on a HEAT N-C arm. Ras binds to the GRD site with Arg1276 being the critical residue for binding. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
| Line 41: | Line 41: | ||
<scene name='90/904311/Closed_arg/4'>Arg1276 in closed conformation </scene> | <scene name='90/904311/Closed_arg/4'>Arg1276 in closed conformation </scene> | ||
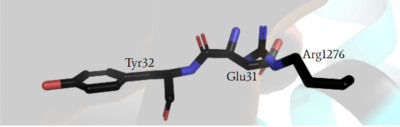
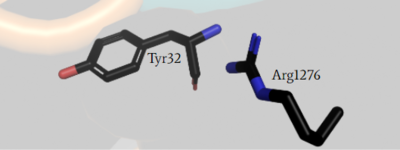
<scene name='90/904312/Closed_zoom/1'>(zoomed in)</scene> has steric clashing with Ras making binding of Ras impossible (Figure #). In the closed confirmation the Arginine finger attempts to bind the backbone gamma carbon(RAS) of a Tyrosine residue(Y32) to assist in hydrolysis, but instead clashes with a Glutamate residue(E31) that would otherwise not interfere in the open conformation and can no longer bind properly. This residue impacts binding of Ras and is of critical importance in determining if Ras is able to bind to the GRD site or not to assist in eventual hydrolysis of GTP. | <scene name='90/904312/Closed_zoom/1'>(zoomed in)</scene> has steric clashing with Ras making binding of Ras impossible (Figure #). In the closed confirmation the Arginine finger attempts to bind the backbone gamma carbon(RAS) of a Tyrosine residue(Y32) to assist in hydrolysis, but instead clashes with a Glutamate residue(E31) that would otherwise not interfere in the open conformation and can no longer bind properly. This residue impacts binding of Ras and is of critical importance in determining if Ras is able to bind to the GRD site or not to assist in eventual hydrolysis of GTP. | ||
| - | Mutations to the arginine finger are shown to slow function of NF1 and other GTPases in their function in accelerating GTP hydrolysis. When a Ras GAP such as NF1 can functionally utilize its arginine finger, it allows NF1 to rapidly hydrolyze GTP when bound to Ras. This emphasizes the importance of the R1276 residue for the GRD site binding to Ras.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> [[Image:ClosedArgFinger.PNG|400px|left|thumb| blah]] [[Image:OpenArgFinger.PNG|400px| | + | Mutations to the arginine finger are shown to slow function of NF1 and other GTPases in their function in accelerating GTP hydrolysis. When a Ras GAP such as NF1 can functionally utilize its arginine finger, it allows NF1 to rapidly hydrolyze GTP when bound to Ras. This emphasizes the importance of the R1276 residue for the GRD site binding to Ras.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| + | [[Image:ClosedArgFinger.PNG|400px|left|thumb| blah]] [[Image:OpenArgFinger.PNG|400px|none|thumb|blah]] | ||
==SPRED 1== | ==SPRED 1== | ||
SPRED 1 is a protein that binds to the GAPex domain of Neurofibromin. Its function recruits the Neurofibromin protein when bound from the cytosol to the plasma membrane. SPRED 1 will bind to the GAPex domain of Neurofibromin in the <scene name='90/904311/Sped-1_closed/1'>closed conformation'</scene> in the cytosol to recruit Neurofibromin to the plasma membrane. Unlike Ras, SPRED-1 does show the ability to <scene name='90/904311/Spred-1_open/1'>bind to the open conformation</scene>. When bound to the open conformation of Neurofibromin in the cytosol, it may present a different orientation that impacts the recruitment to the plasma membrane. Further research is needed to assess the impact of the function and the changes it may present.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | SPRED 1 is a protein that binds to the GAPex domain of Neurofibromin. Its function recruits the Neurofibromin protein when bound from the cytosol to the plasma membrane. SPRED 1 will bind to the GAPex domain of Neurofibromin in the <scene name='90/904311/Sped-1_closed/1'>closed conformation'</scene> in the cytosol to recruit Neurofibromin to the plasma membrane. Unlike Ras, SPRED-1 does show the ability to <scene name='90/904311/Spred-1_open/1'>bind to the open conformation</scene>. When bound to the open conformation of Neurofibromin in the cytosol, it may present a different orientation that impacts the recruitment to the plasma membrane. Further research is needed to assess the impact of the function and the changes it may present.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | ||
Revision as of 13:34, 29 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Neurofibromin 1
| |||||||||||