Sandbox Reserved 1703
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Overall Structure=== | ===Overall Structure=== | ||
[https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM] studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2<ref name="Lin" />. The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Vft/1'>Venus FlyTrap Domain(VFT)</scene>, followed by a <scene name='90/904307/Crds/1'>Cysteine Rich Domain</scene> linker to the Transmembrane Domain that contains <scene name='90/904307/Inactive_7tm/1'>7 alpha helices (7TM)</scene> on both the <scene name='90/904308/Alphaandbetachain/1'>alpha and beta</scene> chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. MGlu2 is a homodimer which is imperative to the receptor’s ability to relay signals induced by glutamate from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM] studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2<ref name="Lin" />. The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Vft/1'>Venus FlyTrap Domain(VFT)</scene>, followed by a <scene name='90/904307/Crds/1'>Cysteine Rich Domain</scene> linker to the Transmembrane Domain that contains <scene name='90/904307/Inactive_7tm/1'>7 alpha helices (7TM)</scene> on both the <scene name='90/904308/Alphaandbetachain/1'>alpha and beta</scene> chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. MGlu2 is a homodimer which is imperative to the receptor’s ability to relay signals induced by glutamate from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | ||
| + | [[Image:IMG 1FBB977FEDDF-1.pdf|400 px|right|thumb|'''Figure 2.'''The demonstrates the transformation of mGlu2 from inactive, to intermediate, to PAM-bound, to fully active.]] | ||
===Inactive State=== | ===Inactive State=== | ||
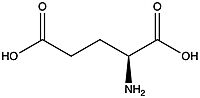
A few hallmarks of the inactive structure of mGlu2 are the Venus FlyTrap Domain in the open conformation, well separated Cysteine-Rich Domains, and distinct orientation of the 7 Transmembrane Domains (7TM). Perhaps the most critical component of the inactive form is the <scene name='90/904307/Tmd_helices/3'>asymmetric TM3-TM4 interface</scene> formed by both of the 7 alpha helices in the alpha and beta chains in the transmembrane domain. The transmembrane domain is mediated mainly by helix IV on the alpha chain and helix lll on the beta chain of the dimer through hydrophobic interactions. These hydrophobic interactions between both transmembrane helices stabilize inactive conformation of mGlu2<ref name="Lin"/>. | A few hallmarks of the inactive structure of mGlu2 are the Venus FlyTrap Domain in the open conformation, well separated Cysteine-Rich Domains, and distinct orientation of the 7 Transmembrane Domains (7TM). Perhaps the most critical component of the inactive form is the <scene name='90/904307/Tmd_helices/3'>asymmetric TM3-TM4 interface</scene> formed by both of the 7 alpha helices in the alpha and beta chains in the transmembrane domain. The transmembrane domain is mediated mainly by helix IV on the alpha chain and helix lll on the beta chain of the dimer through hydrophobic interactions. These hydrophobic interactions between both transmembrane helices stabilize inactive conformation of mGlu2<ref name="Lin"/>. | ||
| - | [[Image:TM4_hydrophobic_interactions.png|300 px|left|thumb|'''Figure | + | [[Image:TM4_hydrophobic_interactions.png|300 px|left|thumb|'''Figure 3.''' Hydrophobic interactions of transmembrane helices III and IV that stabilize the inactive form of mGlu2.]] |
===Intermediate Form=== | ===Intermediate Form=== | ||
Although there are no Cryo-EM images of the intermediate form, it is still a very important state that mGlu2 goes through. The <scene name='90/904308/Agonist_binding_site/4'>Agonist Binding Site</scene> is formed by both lobes of the Venus FlyTrap Domain. The receptor will remain in this inactive state if there are insufficient concentrations of glutamate available<ref name="Du" />. Since glutamate is the main excitatory neurotransmitter in the central nervous system, its ability to bind is extremely important, especially for cell excitability. | Although there are no Cryo-EM images of the intermediate form, it is still a very important state that mGlu2 goes through. The <scene name='90/904308/Agonist_binding_site/4'>Agonist Binding Site</scene> is formed by both lobes of the Venus FlyTrap Domain. The receptor will remain in this inactive state if there are insufficient concentrations of glutamate available<ref name="Du" />. Since glutamate is the main excitatory neurotransmitter in the central nervous system, its ability to bind is extremely important, especially for cell excitability. | ||
| Line 23: | Line 24: | ||
A positive allosteric modulator (PAM) or a negative allosteric modulator (NAM) can bind to mGlu2. <scene name='90/904308/Pam/3'>PAM binds</scene> to the receptor, induces conformational changes, which helps to promote greater affinity for G protein binding. PAM binds in a binding pocket that is created by alpha helices III, V, VI, VII in the transmembrane domain. Upon binding of PAM, it interacts with helix VI, including residues W773, F776, L777, and F780. Due to spatial hindrance, helix VI is shifted downward, causing conformational changes. NAM, however, reduces the affinity for G protein binding. NAM binds to the same binding pocket as PAM and also interacts with residue W773. Due to the structure of NAM, it occupies the binding site a little deeper than PAM. This causes NAM to push on the side chain of W773 towards helix VII<ref name="Lin"/>. PAM and NAM induce different conformational changes, which result in different outcomes. | A positive allosteric modulator (PAM) or a negative allosteric modulator (NAM) can bind to mGlu2. <scene name='90/904308/Pam/3'>PAM binds</scene> to the receptor, induces conformational changes, which helps to promote greater affinity for G protein binding. PAM binds in a binding pocket that is created by alpha helices III, V, VI, VII in the transmembrane domain. Upon binding of PAM, it interacts with helix VI, including residues W773, F776, L777, and F780. Due to spatial hindrance, helix VI is shifted downward, causing conformational changes. NAM, however, reduces the affinity for G protein binding. NAM binds to the same binding pocket as PAM and also interacts with residue W773. Due to the structure of NAM, it occupies the binding site a little deeper than PAM. This causes NAM to push on the side chain of W773 towards helix VII<ref name="Lin"/>. PAM and NAM induce different conformational changes, which result in different outcomes. | ||
| - | [[Image:PAM binding pocket correct.png |300px|right|thumb|'''Figure | + | [[Image:PAM binding pocket correct.png |300px|right|thumb|'''Figure 4.'''This is PAM located in its binding pocket. PAM, JNJ-40411813, is shown in magenta and colored by atom. The image shows four labelled alpha helices (III, V, VI, and VII) that create the binding pocket in the 7TM region of mGlu2 for PAM to bind within. The binding of PAM promotes the function of the mGLu2.]] |
===Active State=== | ===Active State=== | ||
| Line 33: | Line 34: | ||
====G-protein Binding==== | ====G-protein Binding==== | ||
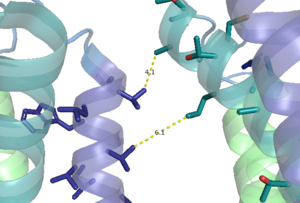
The PAM induced downward shift of helix IV coupled with the reorientation of the transmembrane domain to a TM6-TM6 asymmetric interface, opens up a cleft on the intracellular surface of the receptor. This cleft allows a hook-like region (figure 4), that is composed of the last 4 residues of the alpha subunit of the G-protein, to move in adjacent to helix IV in the transmembrane domain. One very important residue in this interaction is <scene name='90/904308/Binding_recognition_site/5'>C351</scene> on the hook that participates in hydrophobic interactions with Intracellular loop 2 and helix IV. It is due to these interactions that the C-terminal region of the alpha subunit of the G-protein binds in the shallow groove formed by intracellular loops 2 and 3 and residues on helices lll and lV<ref name="Lin" />.The receptor is now <scene name='90/904308/Active_structure/3'>fully active</scene> with the dimer coupled only to one G-protein, the Venus FlyTrap Domain in the closed conformation resulting in a tighter form, and the transmembrane domain helices reoriented on both the alpha and beta chains to form an asymmetric dimer interface. | The PAM induced downward shift of helix IV coupled with the reorientation of the transmembrane domain to a TM6-TM6 asymmetric interface, opens up a cleft on the intracellular surface of the receptor. This cleft allows a hook-like region (figure 4), that is composed of the last 4 residues of the alpha subunit of the G-protein, to move in adjacent to helix IV in the transmembrane domain. One very important residue in this interaction is <scene name='90/904308/Binding_recognition_site/5'>C351</scene> on the hook that participates in hydrophobic interactions with Intracellular loop 2 and helix IV. It is due to these interactions that the C-terminal region of the alpha subunit of the G-protein binds in the shallow groove formed by intracellular loops 2 and 3 and residues on helices lll and lV<ref name="Lin" />.The receptor is now <scene name='90/904308/Active_structure/3'>fully active</scene> with the dimer coupled only to one G-protein, the Venus FlyTrap Domain in the closed conformation resulting in a tighter form, and the transmembrane domain helices reoriented on both the alpha and beta chains to form an asymmetric dimer interface. | ||
| - | [[Image:Newly labled hook region.png|400 px|left|thumb|'''Figure | + | [[Image:Newly labled hook region.png|400 px|left|thumb|'''Figure 5.''' The hook-like region is made up of the last 4 residues on the alpha subunit of the G-protein. Residue C351 hydrophobically interacts with intracellular loop 2 and helix IV. Due to these interactions, the G-protein is able to bind to a shallow groove formed by intracellular loops 2 and 3.]] |
==Clinical Relevance== | ==Clinical Relevance== | ||
Revision as of 14:41, 29 March 2022
==Metabotropic Glutamate Receptor 2==
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Lin S, Han S, Cai X, Tan Q, Zhou K, Wang D, Wang X, Du J, Yi C, Chu X, Dai A, Zhou Y, Chen Y, Zhou Y, Liu H, Liu J, Yang D, Wang MW, Zhao Q, Wu B. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021 Jun;594(7864):583-588. doi: 10.1038/s41586-021-03495-2. Epub 2021, Jun 16. PMID:34135510 doi:http://dx.doi.org/10.1038/s41586-021-03495-2
- ↑ Seven, Alpay B., et al. “G-Protein Activation by a Metabotropic Glutamate Receptor.” Nature News, Nature Publishing Group, 30 June 2021, https://www.nature.com/articles/s1586-021-03680-3
- ↑ Zhang, Zhu, et al. “Roles of Glutamate Receptors in Parkinson's Disease.” MDPI, Multidisciplinary Digital Publishing Institute, 6 Sept. 2019, https://dx.doi.org/10.3390%2Fijms20184391.>
- ↑ Yang, Hong-Ju, et al. “Deletion of Type 2 Metabotropic Glutamate Receptor Decreases Sensitivity to Cocaine Reward in Rats.” Cell Reports, U.S. National Library of Medicine, 11 July 2017, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5555082/.>
- ↑ 5.0 5.1 Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.>
- ↑ Ellaithy A, Younkin J, Gonzalez-Maeso J, Logothetis DE. Positive allosteric modulators of metabotropic glutamate 2 receptors in schizophrenia treatment. Trends Neurosci. 2015 Aug;38(8):506-16. doi: 10.1016/j.tins.2015.06.002. Epub, 2015 Jul 4. PMID:26148747 doi:http://dx.doi.org/10.1016/j.tins.2015.06.002
- ↑ Muguruza C, Meana JJ, Callado LF. Group II Metabotropic Glutamate Receptors as Targets for Novel Antipsychotic Drugs. Front Pharmacol. 2016 May 20;7:130. doi: 10.3389/fphar.2016.00130. eCollection, 2016. PMID:27242534 doi:http://dx.doi.org/10.3389/fphar.2016.00130
Student Contributors
Frannie Brewer and Ashley Wilkinson