This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
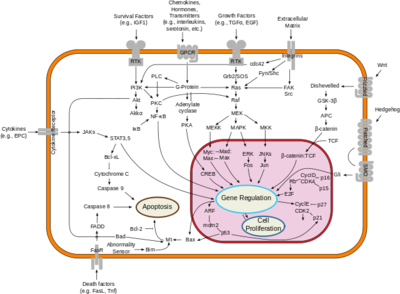
Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1 (NF1). This condition drives several forms of human cancers by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people, and the NF gene itself has the highest mutation rate of any known human gene, adding to its prevalence<ref name= ''Lupton''>PMID:34887559</ref>. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems<ref name= ''Cimino''>PMID:29478615</ref>. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein<ref name= ''Ly''>PMID:31582003</ref>. | Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1 (NF1). This condition drives several forms of human cancers by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people, and the NF gene itself has the highest mutation rate of any known human gene, adding to its prevalence<ref name= ''Lupton''>PMID:34887559</ref>. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems<ref name= ''Cimino''>PMID:29478615</ref>. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein<ref name= ''Ly''>PMID:31582003</ref>. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| + | <scene name='90/904326/Open_conformation/2'>Open Conformation</scene>, | ||
| + | <scene name='90/904326/Open_conformation_with_grd_hig/2'>Open Conformation with GRD highlighted</scene> | ||
| + | <scene name='90/904326/Closed_conformation/2'>Closed Conformation</scene>, | ||
| + | <scene name='90/904326/Grd_closed_conformation/2'>Closed Conformation with GRD highlighted</scene> | ||
| + | <scene name='90/904326/Sec15ph_and_grd_open/2'>sec14ph domain in Open Conformation</scene> | ||
| + | <scene name='90/904326/Sec14ph_and_grd_closed/2'>sec14ph domain in Closed Conformation</scene> | ||
| + | <scene name='90/904326/Active_site_with_residues/6'>active site</scene> | ||
</StructureSection> | </StructureSection> | ||
Revision as of 18:32, 31 March 2022
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12138-42. Epub 2002 Sep 4. PMID:12213964 doi:http://dx.doi.org/10.1073/pnas.192453199
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Scheffzek K, Shivalingaiah G. Ras-Specific GTPase-Activating Proteins-Structures, Mechanisms, and Interactions. Cold Spring Harb Perspect Med. 2019 Mar 1;9(3). pii: cshperspect.a031500. doi:, 10.1101/cshperspect.a031500. PMID:30104198 doi:http://dx.doi.org/10.1101/cshperspect.a031500
- ↑ Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3649-53. doi: 10.1073/pnas.89.8.3649. PMID:1565661 doi:http://dx.doi.org/10.1073/pnas.89.8.3649
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky