We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
The <scene name='90/904326/Active_site_with_residues/6'>active site</scene> for GTP hydrolysis of Ras is located in the Gap-related domain of neurofibromin. The catalytic residues include R68, Q61, and Y32, as well as magnesium and water molecules. Arginine is referred to as an “arginine finger” because it points into the binding site of GTP to stabilize and orient the position of glutamine through a network of hydrogen bonds between water molecules. This arginine comes from the Gap-related domain of neurofibromin. When GDP is bound, glutamine is too far away to perform its catalytic action. Glutamine interacts with the gamma phosphate via a hydrogen bond created from an interaction between a water molecule and the gamma phosphate. When GTP is bound, tyrosine moves inward to face it. In the GDP bound form, tyrosine faces outward. | The <scene name='90/904326/Active_site_with_residues/6'>active site</scene> for GTP hydrolysis of Ras is located in the Gap-related domain of neurofibromin. The catalytic residues include R68, Q61, and Y32, as well as magnesium and water molecules. Arginine is referred to as an “arginine finger” because it points into the binding site of GTP to stabilize and orient the position of glutamine through a network of hydrogen bonds between water molecules. This arginine comes from the Gap-related domain of neurofibromin. When GDP is bound, glutamine is too far away to perform its catalytic action. Glutamine interacts with the gamma phosphate via a hydrogen bond created from an interaction between a water molecule and the gamma phosphate. When GTP is bound, tyrosine moves inward to face it. In the GDP bound form, tyrosine faces outward. | ||
====Arginine Finger==== | ====Arginine Finger==== | ||
| - | + | ||
| - | + | ||
====Conformations==== | ====Conformations==== | ||
Neurofibromin is a dimeric protein that is found in the cytoplasm. It exists in two conformations, <scene name='90/904326/Open_conformation/2'>open</scene> and <scene name='90/904326/Closed_conformation/2'>closed</scene> . The open conformation has one of the protomers in an auto-inhibited conformation and the other in an open conformation. In the <scene name='90/904326/Open_conformation_with_grd_hig/2'>open conformation</scene>, Ras is able to bind to the GRD neurofibromin. The <scene name='90/904326/Grd_closed_conformation/2'>closed conformation</scene> has both protomers in an autoinhibited conformation, which sterically hinders the binding of Ras to GRD. | Neurofibromin is a dimeric protein that is found in the cytoplasm. It exists in two conformations, <scene name='90/904326/Open_conformation/2'>open</scene> and <scene name='90/904326/Closed_conformation/2'>closed</scene> . The open conformation has one of the protomers in an auto-inhibited conformation and the other in an open conformation. In the <scene name='90/904326/Open_conformation_with_grd_hig/2'>open conformation</scene>, Ras is able to bind to the GRD neurofibromin. The <scene name='90/904326/Grd_closed_conformation/2'>closed conformation</scene> has both protomers in an autoinhibited conformation, which sterically hinders the binding of Ras to GRD. | ||
| Line 23: | Line 22: | ||
<scene name='90/904325/Ras_full_structure/2'>Full Ras protein with GTP ligand</scene> | <scene name='90/904325/Ras_full_structure/2'>Full Ras protein with GTP ligand</scene> | ||
===Mechanism of Ras Coupled with Neurofibromin=== | ===Mechanism of Ras Coupled with Neurofibromin=== | ||
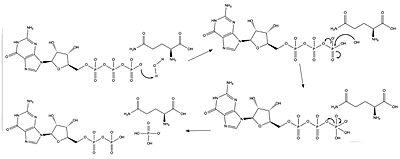
| - | The RasGAP interactions that occur when neurofibromin's GAP domain and Ras are bound have two critical catalytic components. The first is the <scene name='90/904325/Arginine_finger_and_gtp/1'>the arginine finger</scene> of the NF protein, which stabilizes the catalytic glutamine, as well as stabilizing the transition state of the phosphoryl transfer<ref name= ''Scheffzek''>PMID:30104198</ref>. The second component is the catalytic glutamine of the Ras protein, which stabilizes the nucleophilic water as it attacks the third phosphate group in the GTP molecule as shown in Figure 1<ref name= ''Prive''>PMID:1565661</ref>. | + | The RasGAP interactions that occur when neurofibromin's GAP domain and Ras are bound have two critical catalytic components. The first is the <scene name='90/904325/Arginine_finger_and_gtp/1'>the arginine finger</scene> of the NF protein, which stabilizes the catalytic glutamine, as well as stabilizing the transition state of the phosphoryl transfer<ref name= ''Scheffzek''>PMID:30104198</ref>. The second component is the <scene name='90/904325/Arginine_finger_and_gdp/2'> catalytic glutamine </scene> of the Ras protein, which stabilizes the nucleophilic water as it attacks the third phosphate group in the GTP molecule as shown in Figure 1<ref name= ''Prive''>PMID:1565661</ref>. |
[[Image:Ras mechanism.jpg|400 px|right|thumb|Figure 1: The catalytic glutamine stabilizes the nucleophilic water.]] | [[Image:Ras mechanism.jpg|400 px|right|thumb|Figure 1: The catalytic glutamine stabilizes the nucleophilic water.]] | ||
===Downstream Effects=== | ===Downstream Effects=== | ||
Revision as of 18:54, 31 March 2022
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12138-42. Epub 2002 Sep 4. PMID:12213964 doi:http://dx.doi.org/10.1073/pnas.192453199
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Scheffzek K, Shivalingaiah G. Ras-Specific GTPase-Activating Proteins-Structures, Mechanisms, and Interactions. Cold Spring Harb Perspect Med. 2019 Mar 1;9(3). pii: cshperspect.a031500. doi:, 10.1101/cshperspect.a031500. PMID:30104198 doi:http://dx.doi.org/10.1101/cshperspect.a031500
- ↑ Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3649-53. doi: 10.1073/pnas.89.8.3649. PMID:1565661 doi:http://dx.doi.org/10.1073/pnas.89.8.3649
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky