We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1702
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Structure == | == Structure == | ||
===Overall Structure=== | ===Overall Structure=== | ||
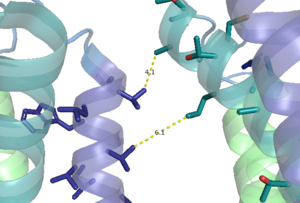
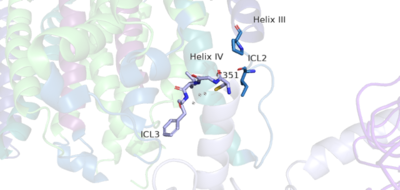
| - | Cryo-EM studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2 (Lin). The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Better_inactive_structure/3'>Venus Fly Trap Domain (VFT)</scene>, followed by a <scene name='90/904307/Better_inactive_structure/2'>Cysteine Rich Domain</scene> linker to the | + | Cryo-EM studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2 (Lin). The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Better_inactive_structure/3'>Venus Fly Trap Domain (VFT)</scene>, followed by a <scene name='90/904307/Better_inactive_structure/2'>Cysteine Rich Domain</scene> linker to the <scene name='90/904307/Better_inactive_structure/4'>Transmembrane Domain</scene> that contains 7 alpha helices (7TM) on both the alpha and beta chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. MGlu2 is a homodimer which is imperative to the receptor’s ability to relay signals induced by glutamate from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2. |
===Inactive=== | ===Inactive=== | ||
Revision as of 14:45, 5 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Metabotropic Glutamate Receptor 2
| |||||||||||