T. brucei Phosphodiesterase B1
From Proteopedia
| Line 17: | Line 17: | ||
== Structural highlights == | == Structural highlights == | ||
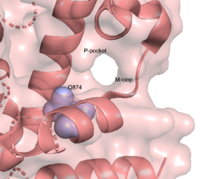
| - | There are several resolved crystal structures for TbPDEB1 in both apo and holo forms which facilitate virtual screenings of libraries of compounds for inhibitory activity against TbPDEB1. The resolved crystal structure for the catalytic domain of TbPDEB1 (PDB identifier: 4I15) confirmed the existence of a ''T. brucei'' specific '''P-pocket''', a new and exciting direction for improving drug selectivity for ''T. brucei'' phosphodiesterases <ref>PMID:23409953</ref>. | + | There are several resolved crystal structures for TbPDEB1 in both apo and holo forms which facilitate virtual screenings of libraries of compounds for inhibitory activity against TbPDEB1. The resolved crystal structure for the catalytic domain of TbPDEB1 (PDB identifier: 4I15) confirmed the existence of a ''T. brucei'' specific '''P-pocket''', a new and exciting direction for improving drug selectivity for ''T. brucei'' phosphodiesterases. The active site for TbPDEB1 contains Magnesium and Zinc ions with each metal ion forming an octahedral geometry connecting to catalytic histidine and aspartic residues and water molecules.<ref>PMID:23409953</ref>. |
| - | { | + | { |
| [[Image:CatalyticDomain2.png|left|200px]] | | [[Image:CatalyticDomain2.png|left|200px]] | ||
| [[Image:Ppocket2.png|left|200px]] | | [[Image:Ppocket2.png|left|200px]] | ||
| - | | | + | | |
| - | + | } | |
| - | Only the catalytic domain of the TbPDEB1 enzyme performs | + | Only the catalytic domain of the TbPDEB1 enzyme performs hydrolysis of cAMP to AMP <ref>PMID:19513125</ref>. |
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| - | |||
| - | </StructureSection> | ||
| - | |||
== Citations == | == Citations == | ||
| - | |||
| - | <references/> | ||
Revision as of 19:13, 12 April 2022
T. brucei PhosphodiesteraseB1
|
Contents |
Function
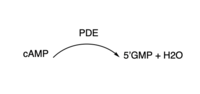
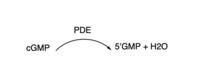
3′,5′-cyclic nucleotide phosphodiesterases (PDEs) are enzymes that hydrolyse 3',5'-phosphodiester bonds in two of the most important cell signalling molecules, cyclic-adenosine monophosphates (cAMPs) and cyclic-guanosine monophosphates (cGMPs). PDEs are essential for the inactivation of cAMP and or cGMP to regulate their intracellular concentrations and maintain cellular homeostasis [1]. PDEs directly modulate these messenger molecules by degrading cAMP or cGMP when concentrations are elevated beyond the cellular threshold [2]. is one of two cyclic nucleotide phosphodiesterases (the other is TbrPDEB2) that have been validated as promising drug targets for the treatment of Human African Trypanosomiasis (HAT), a life-threatening infectious disease also known as sleeping sickness which is caused by Trypanosoma brucei parasites [3]. PDEB1 is a cAMP-specific hydrolase that catalyses the hydrolysis of cAMP to AMP in the catalytic domain. This enzyme has been reported to play a major role in facilitating Trypanosoma motility in host cells and subsequent progression through the parasite life cycle [4].
Relevance
Trypanosomiasis, or sleeping sickness, is a neglected tropical disease that is fatal if left untreated and is one of the major causes of human and livestock mortality in developing countries. This infection also affects about 72% of European and American tourists who visit safari reserves and game parks in endemic regions [5]. The current treatment options for trypanosomiasis are outdated - such as melarsoprol which was first introduced in 1949- and also sub-optimal typically showing high toxicity, particularly in late-stage infections [6]. There is a clear need for safer, more efficacious drugs to treat sleeping sickness which is why creative studies are being performed to validate new drug targets for trypanosomiasis drug development. PDEB1 makes an interesting drug target for HAT drug discovery due to its essential functions that contribute to parasite fitness and survival.
Structural highlights
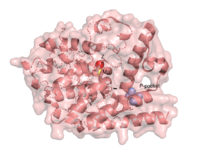
There are several resolved crystal structures for TbPDEB1 in both apo and holo forms which facilitate virtual screenings of libraries of compounds for inhibitory activity against TbPDEB1. The resolved crystal structure for the catalytic domain of TbPDEB1 (PDB identifier: 4I15) confirmed the existence of a T. brucei specific P-pocket, a new and exciting direction for improving drug selectivity for T. brucei phosphodiesterases. The active site for TbPDEB1 contains Magnesium and Zinc ions with each metal ion forming an octahedral geometry connecting to catalytic histidine and aspartic residues and water molecules.[7]. {
| || } Only the catalytic domain of the TbPDEB1 enzyme performs hydrolysis of cAMP to AMP [8]. This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.