Introduction
Metabotropic glutamate receptors are found in the central nervous system and play a critical role in modulating cell excitability and synaptic transmission
[1].
Glutamate, shown in Figure 1, is a negatively charged polar amino acid that is the main neurotransmitter in the brain. Glutamate activates 8 different types of metabotropic glutamate receptors
[2].
Metabotropic Glutamate Receptor 2 (mGlu2) is a member of the
Class C GPCRFamily and can further be classified into the Group II subgroup of metabotropic receptors. Since mGlu2 is a part of the Class C GPCR family, it undergoes small conformational changes to the transmembrane domain (TMD) to move from the inactive to the fully active structure. Class A and B GPCR Families, however, have a different mechanism that causes conformational changes to Transmembrane 6
[2]. mGlu2 functionality is dependent on the concentration of glutamate where higher concentrations of glutamate will promote stronger signal transduction from the extracellular domain to the transmembrane domain
[1].
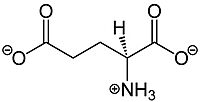
Figure 1.The structure of glutamate that activates mGlu2.
mGlu2 plays vital roles in memory formation, pain management, and addiction, which makes it an important drug target for Parkinson’s Disease,Schizophrenia, cocaine dependence, and many other neurological conditions.
Structure
Overall Structure
Cryo-EM studies of mGlu2 have yielded adequate structural maps of mGlu2 in various activation states. These maps provided clearer understanding of the conformational changes between the inactive and active states of mGlu2[1]. The overall of the mGlu2 is composed of 3 main parts: a ligand binding , followed by a linker to the Transmembrane Domain that contains on both the chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers.
mGlu2 is a homodimer. Dimerization of mGlu2 is required to relay glutamate binding from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2[3]. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2[1].
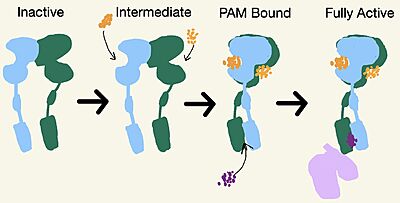
Figure 2. Demonstrates the conformational changes of mGlu2.
Inactive State
A few hallmarks of the inactive structure of mGlu2 are the Venus FlyTrap Domain in the open conformation, well separated Cysteine-Rich Domains, and distinct orientation of the 7 Transmembrane Domains (7TM). The most critical component of the inactive form is the formed by the 7 α-helices in the α and β chains of the 7TM. The inactive structure of mGlu2 is mediated mainly by helices 3 and 4 on both the α and β chains of the dimer through hydrophobic interactions. These hydrophobic interactions between both transmembrane helices stabilize inactive conformation of mGlu2[1]. Key hydrophobic interactions are between A630 V699 on the helix
Intermediate Form
No Cryo-EM structures are currently available of the intermediate form, but it is still an important state for the full activation of mGlu2. The is formed by both lobes of the Venus FlyTrap Domain. The receptor will remain in this inactive state if there are insufficient concentrations of glutamate available[3]. Since glutamate is the main excitatory neurotransmitter in the central nervous system, its binding controls cell excitability.
PAM and NAM Bound Form
A positive allosteric modulator (PAM) or a negative allosteric modulator (NAM) can bind to mGlu2. PAM and NAM induce different conformational changes, which result in different outcomes. to the receptor, induces conformational changes, which helps to promote greater affinity for G protein binding. PAM binds in a binding pocket that is created by helices III, V, VI, VII in the TMD. Within helix VI, the hydrophobic is is composed of W773, F776, L777, and F780. Due to spatial hindrance, helix VI is shifted downward, causing conformational changes. NAM, however, reduces the affinity for G protein binding. NAM binds to the same binding pocket as PAM and also interacts with residue W773, but NAM occupies the binding site a little deeper than PAM. This causes NAM to push the side chain of W773 towards helix VII[1].
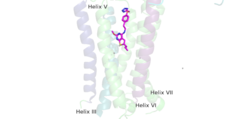
Figure 4.This is PAM located in its binding pocket. PAM, JNJ-40411813, is shown in magenta and colored by atom. The image shows four labelled alpha helices (III, V, VI, and VII) that create the binding pocket in the 7TM region of mGlu2 for PAM to bind within. The binding of PAM promotes the function of the mGLu2.
Active State
Upon binding of the PAM, helix VI is shifted downward in the transmembrane domain. This downward shift induces a reorientation of the transmembrane domain from its original TM3-TM4 asymmetric dimer interface in the inactive form to now a . The downward shift of helix VI is crucial for the receptor’s transformation from the inactive to the active form for 2 main reasons: (1) reorientation breaks key interactions in the transmembrane domain that stabilize the inactive form (2) positions of the helices in the transmembrane domain to assist in the binding and recognitions of the G-Protein. The G-protein is made up of three subunits: , , and a Γ-subunit.
G-Protein Recognition
Reorientation positions helix lll on either the alpha or beta chain because both have the ability to bind to the G-protein but only one chain is required for full receptor activation. The intracellular region of helix lll mainly contributes to the interactions with the alpha subunit of the G-protein. Intracellular Loop 2 plays a key role in G-protein coupling as well by building polar interaction networks through its ionic interactions with the of the G-protein. Lastly, mGlu2 residue E666 forms a salt bridge with an alpha N residue (R32) on the alpha subunit which further destabilizes the inactive conformation[1].
G-protein Binding
The PAM induced downward shift of helix IV coupled with the reorientation of the transmembrane domain to a TM6-TM6 asymmetric interface, opens up a cleft on the intracellular surface of the receptor. This cleft allows a hook-like region (figure 4), from the last 4 terminals of the α-subunit of the G-protein, to move in adjacent to helix IV in the transmembrane domain. Within this interaction, on the hook participates in hydrophobic interactions with Intracellular loop 2 and helix 4. These interactions allow the C-terminal region of the G-protein α-subunit to bind in the shallow groove formed by intracellular loops 2 and 3 and residues on helices 3 and lV[1].The receptor is now with the dimer coupled only to one G-protein. The Venus FlyTrap Domain is in the closed conformation resulting in a tighter form. The TMD helices are also reoriented in both monomers to form an asymmetric dimer interface.
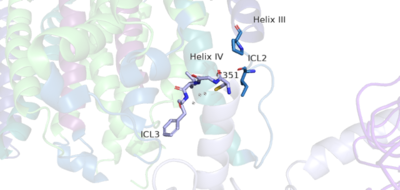
Figure 5. The hook-like region is made up of the last 4 residues on the alpha subunit of the G-protein. Residue C351 hydrophobically interacts with intracellular loop 2 and helix IV. Due to these interactions, the G-protein is able to bind to a shallow groove formed by intracellular loops 2 and 3.
Clinical Relevance
Metabotropic Glutamate Receptors play a variety of roles both presynaptically and postsynaptically in the Central Nervous System. Misregulation of metabotropic glutamate receptors can cause certain types of diseases, while manipulation of these receptors are starting to be used as drug targets.
Schizophrenia
Schizophreniais a chronic brain disorder that affects a person’s ability to think, feel, and behave clearly. The exact cause of Schizophrenia is unknown currently[4]. The symptoms from the disease can vary from patient to patient, but they can be broken down into positive, negative, and cognitive symptoms[4]. Although antipsychotic drugs help to treat Schizophrenia, these drugs only target positive symptoms and have limited efficacy against negative and cognitive symptoms [4]. mGlu2 receptors are a therapeutic target for Schizophrenia, as mGlu2 receptors are expressed in regions associated with Schizophrenia, such as the prefrontal cortex, hippocampus, the thalamus, and amygdala [5]. Specifically, mGlu2 agonist, PAM, exhibits antipsychotic properties by increasing dopamine extracellular levels. Increasing dopamine levels improves negative symptoms of Schizophrenia. mGlu2 agonists also increase cortical serotonin levels, which is a property seen in many antipsychotic drugs. These clinical properties give potential for mGlu2 and PAM as future treatments for Schizophrenia[4].




