MRGPRX2 is a certain type of GPCR that is located in the cellular membranes of mast cells.
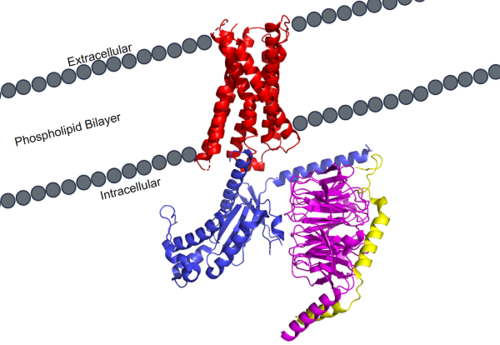
Figure 1. MRGPRX2 as it sits within the cellular membrane. Phospholipid bilayer is represented by grey dots, with labeled cellular locations
Background
GCPR’s or G-Protein Coupled Receptors are a special type of protein receptor that promotes cellular signaling. Due to its structure spanning the cellular membrane, it works to transmit extracellular signals to create a change inside the cell. This is called signal transduction, and is a common way extracellular signals produce a change within cells. Some common cellular pathways that utilize GPCR’s are found in Rhodopsin, a protein essential for the human vision response, or the adrenaline fight-or-flight response. Understanding GPCR’s and how they produce their desired intracellular signal is essential understanding essential cellular pathways, especially in their diseased states. As of 2017, there were about 475 drugs in circulation that acted on 108 GPCR targets, and at least 321 GPCR-targeting drugs were in clinical trial stages, with about 20% of them targeting novel GPCR’s. [1] Because of the clinical relevance of GPCR’s, every new structure found, such as MRGPRX2, contributes to essential drug development to both treat disease, or modulate harmful side effects.
Because of how many different types of GPCR’s there are, they have been categorized into 6 different classes based on shared sequences and functions.[2] MRGPRX2 is categorized into the Class A receptor family, however it has important differences that make it a unique type of Class A receptor. [3][4]
GPCR Structure
Cryo-electron microscopy (cryo-EM) was used to image the MRGPRX2 receptor to analyze its structure. [3] [4] which helped to classify it into the A family of GPCR's. MRGPRX2 therefore shares the same general structural domains of all GPCR's. This includes a and a domain. The G-protein domain consists of , , and subunits. In preparing the protein sample, MRGPRX2 was prepared with the in order to stabilize the membrane for proper imaging. For simplicity and to focus on the specific MRGPRX2 receptor, the antibody has been removed.
Transmembrane Domain
The transmembrane domain spans the cellular membrane. It consists of and (three extracellular loops, and three intracellular loops). The transmembrane helices are numbered 1-7 and contain special conserved motifs that are shared across other A family receptors. These motifs are expanded upon later, as they heavily contribute to the structure and therefore function of the transmembrane domain as a whole.
The extracellular domain of the protein is responsible for ligand binding, which initiates signal transduction. The properties of the extracellular environment of the transmembrane domain determines what ligands bind to the protein, and what type of binding interactions the protein and ligand have. In MRGPRX2, the extracellular side of the protein has one binding pocket with . Sub-pocket 1 is negatively charged due to negatively charged residues (D184 and E164), while sub-pocket 2 contains hydrophobic amino acids which contribute to hydrophobic interactions between the ligand and protein.
The intracellular domain is what connects the transmembrane and G-protein domains together. There are a wide variety of residues and important interactions that contribute to this interaction, and it is very important in transmitting the extracellular signal of ligand binding to the intracellular environment where the G-protein binds and can become activated.
G-Protein
GTP-binding proteins, also known as G-proteins, are heterotrimeric complexes consisting of alpha, beta, and gamma subunits that interact with membrane receptor proteins. G-proteins are responsible transmitting extracellular signals into the cell upon activation. This activation happens when the alpha subunit of the G-protein binds GTP instead of GDP, and then disassociates from the rest of the protein, initiating the intracellular signaling cascade. There are different families of G-alpha subunits, Gαi, Gαs, Gα12/13, and Gαq [5]. MRGPRX2 has been found to bind both Gαi and Gαq subunits with relatively no major structural changes between the two despite slightly different amino acids present [3] [4]. Throughout this page, MGPRX2 is always shown with Gq. Figure 2a shows the overlay of MGPRX2 with either the Gq or Gi alpha subunit. Figure 2b shows the specific residues involved in the between the membrane receptor and G protein. The major difference between the two comes from one amino acid difference (valine on Gq versus phenylalanine on Gi) that pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein. This is the only major structural difference between Gq and Gi subunits.
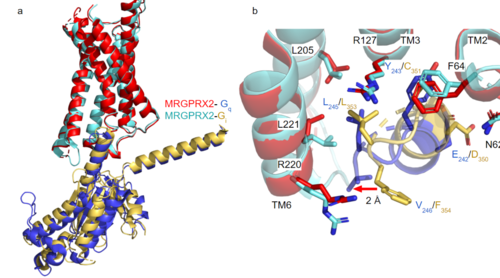
Figure 2a. Overlay of MGPRX2-Gq (red-dark blue) and MGPRX2-Gi (cyan-yellow).
Figure 2b. Important residues involved in the interface between MGPRX2 and Gq/ Gi subunits. Arrow pointing to the major difference between the interfaces, which comes from the final C-terminus residue on the G-alpha subunit. In Gq, there is a valine while in Gi, there is a phenylalanine. This pushes the Gi subunit 2Å away from the arginine residue on helix 6 of the transmembrane protein.
Novel Characteristics
The MRGPRX2 receptor shows surprising differences between it and all other previously characterized class A GPCRs including many conserved class A structural motifs which are absent on MRGPRX2. These structural motif differences contribute to a ligand binding site closer to the membrane surface for MRGPRX2 rather than a ligand binding site deep within the helices (Figure 3). To demonstrate this difference, depth of binding is illustrated by a comparison of MRGPRX2 and 5-HT2AR, another class A GPCR with more conserved structural motifs.
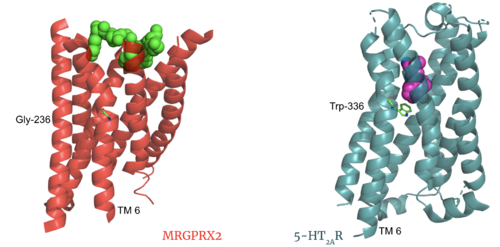
Figure 3. Comparison of ligand Cortistatin-14 binding in MRGPRX2 (left) and binding in 5HT2AR (right)
Toggle Switch
Key toggle switch residues in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off". Toggle switches initiate the transmission of the molecular signal through the 7 TMD helices to the intracellular G protein. Trp-336 is the "iconic" toggle switch in class A GPCR’s [6], and is a part of another motif, known as the CWxP motif. However in MRGPRX2, this tryptophan has been replaced with a [3] [4]. This shift leads to a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helices, especially helix 7 on which the toggle switch is found, pack more tightly. The ligands that interact with MRGPRX2 are able to bind much of the receptor, as opposed to deeper within the helices (Figure 3). This shallower binding pocket expands the types of ligands that are able to interact with X2 and therefore what types of molecules can activate the Human Itch GPCR. More details about what kinds of ligands bind to this receptor are discussed later.
Sodium Site
The allosteric sodium site in class A GPCRs has been characterized as important in inactive state GPCR stabilization [7]. Katritch et al [7] describe that class A GPCRs lacking conserved D2.50 and other polar residues within the sodium pocket are typically inactive. The MRGPRX2 consists of conserved D2.50, or ASP-75, and GLY-116 compared to the previously conserved polar residues in this binding pocket such as S3.39. Other class A GPCRs demonstrate a larger sodium binding pocket with a higher negative character allowing for a suitable environment for sodium ions to bind. In MRGPRX2, this sodium binding pocket lacks the same amount of with the shift to a glycine residue rather than serine. However, evidence suggests that sodium is still able to bind in X2's sodium binding site even with fewer conserved residues.
PIF/LLF Motif
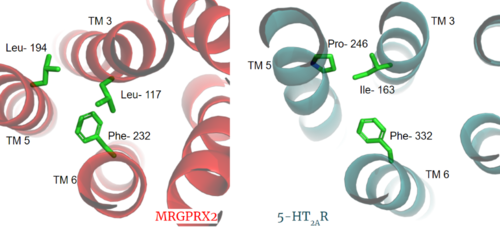
Figure 3. Conserved PIF motif in 5HT2AR (teal) compared to the LLF motif found in MRGPRX2 (red). Transmembrane helices and residues are numbered and labeled to show how this structural change shifts the orientation of the helices.
Another motif found in most, but not all, A family GPCR’s is the PIF motif. The PIF residues () are found on transmembrane helices 5, 3, and 6, respectively. In MRGPRX2, the PIF motif is changed to LLF residues. Figure 3 shows the conserved PIF motif on 5HT2AR, compared to the LLF motif on MRGPRX2. This change to LLF shifts helix 6 towards helix 3, and contributes to the tighter packing of helices [3] [4] and therefore a more surface-level ligand binding site.
DRY/ ERC Motif
The E/DRY motif in most class A GPCRs is responsible for forming salt bridges with surrounding residues and TM6[8]. These salt bridges maintain the inactive conformation of the receptor until ligand binding breaks the ionic "lock" from these interactions. MRGPRX2 has an rather than the typically conserved E/DRY Motif. The amino acid residue shift from TYR-174 to CYS-128 allows compaction of the helices in MRGPRX2 where the standard TYR physically pushes the TMD helices apart(Figure 4). The conserved residues E and R still form salt bridges with nearby residues. This and the closer packing of the helices contribute to a less significant TMD conformational change upon ligand binding (Figure 10).
Disulfide Bonds

Figure 5. Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.
In a large majority of class A GPCRs, there is a conserved disulfide bond between extracellular loop 2 (ECL2) and transmembrane helix 3 (TM3). This bond has been proposed to have a role in structural stability, expression, and function of GPCRs[9]. The MRGPRX2 disulfide bond is between on TM helices 5 and 4, respectively. For example, the shows this disulfide bond between the ECL2 and TM3. Although this bond is in a different location than other class A GPCRs, there is evidence to suggest its location is essential for the signaling of the X2 receptor as the ECL2 instead located at the top of TM4 and TM5 allowing for the large, extracellular binding pocket observed in X2[3].
Further Information
The MRGPRX2 GPCR also contains semi-conserved or fully-conserved motifs that are seen in other class A GPCRs.
NPxxY Motif
The residues in the are pivotal for receptor activation in all Class A GPCRs. This motif is conserved in the MRGPRX2 receptor with residues VAL-231, ASP-75, ASN-275, and TYR-279.
CWxP Motif
The is almost fully conserved except for TRP-236, or the toggle switch, which is replaced with GLY-236. CYS-235, LEU-237, and PRO-238 are all conserved. CYS6.47 may play a fundamental role in GPCR activity by participating in the rearrangement of TM6 and TM7 upon receptor activation[10]. This residue's role is supported by its conservation in MRGPRX2 as it is a functional GPCR.
Ligands
MRGPRX2 binds a wide range of small molecule and peptide ligands. These ligands have positively charged regions which can interact with the negative binding pocket, sub-pocket 1. Some of these larger ligands also have a hydrophobic region which can interact with sub-pocket 2. MRGPRX2 has been known to interact with PAMP-12, Cortistatin-14, C48/80, and Zinc-3573[4],[3].
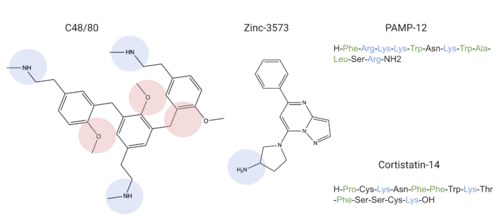
Common MRGPRX2 ligands with positive regions in blue, negative regions in red, and hydrophobic regions in green
Ligands' charge or hydrophilicity are shown to demonstrate where they may interact with sub-pockets 1 and 2. , specifically, interacts with sub-pocket 1 through LYS-3 and sub-pocket 2 through LYS-8.
Function
GPCRs undergo a conformational change in their 7TMD region upon ligand binding. This signal is then transduced to the G-protein allowing for downstream responses due to between the alpha subunit of the G-protein and the transmembrane protein which activates g-protein by GTP exchange.
Before Activation
The culmination of different motifs observed in MRGPRX2 compared to other class A GPCRs leads to external membrane ligand binding. The MRGPRX2 GPCR undergoes a much smaller conformational change upon ligand binding compared to other Class A GPCRs due to surface level binding versus deep helix binding (Figure 10).

Figure 10. Overlay of unbound (transparent) and bound (opaque) transmembrane proteins of both MRGRPX2 (left) and 5-HT2AR (right).
After Activation
After ligand binding and transmembrane protein activation, this signal is transmitted to the alpha subunit of the G-protein which undergoes chemical and conformational changes. The alpha subunit is initially bound with GDP which is then physically replaced by GTP leading to conformational changes. These changes can be seen in this video of a G-protein alpha subunit activation derived from common rats.
Clinical Relevance

Figure 11.Schematic representation of cellular response
Some cells in the human body that express the MRGPRX2 receptor include mast cells in the skin, intestines, and trachea [11]. Mast cells are involved in allergic responses by their release of histamine or other inflammatory chemicals in the body. [12] These responses induce common allergic reaction and anaphylaxis symptoms, such as cutaneous itching sensations or airway constriction
[3][4][12]. Mast cells can be activated by either antibodies in the human immune response or upon ligands binding to their MRGPRX2 receptors [12]. Ligands that bind to MRGPRX2 in the natural environment to produce an allergic response include some contents of insect venom, molecules like C48/80 or other polycationic molecules. They can also respond to endogenous signaling molecules involved in inflammation pathways and immune responses such as cytokines, anaphylatoxins, or neuropeptides [11]. Binding to MRGPRX2 triggers an intracellular signaling pathway that eventually leads to mast cells releasing their contents containing histamine, cytokines, or other inflammatory molecules that may go on to trigger the itching sensation [3] [4].
Many medications commonly list chronic skin itching or inflammatory responses as side effects, such as nateglinide, an anti-diabetic drug [3]. Upon analysis of some medications that list itching as side effects such as nateglinide, they structurally share many similarities to known MRGPRX2 ligands, and could therefore initiate an unwanted itch response in patients. Other drugs such as morphine or codeine also share structural features to known MRGPRX2 ligands, which can contribute to itching side effects seen in users of these drugs [3].
Because of how some drugs initiate an unwanted itch response,
MRGPRX2 has been identified as a potential target for drugs either involved in the inflammatory response or to mediate drugs from causing an unwanted reaction such as Atracurium, Rocuronium, Ciprofloxacin, and Levofloxacin[13]. These drugs have similarly charged regions which may interact with sub-pockets 1 and/or 2. By binding these target drugs, MRGPRX2 can elicit or prevent a response to aid with itch-related side effects.
3D Structures
7s8l, MRGPRX2 Gq
7s8m, MRGPRX2 Gi
6wha, 5HT2AR









