We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1712
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
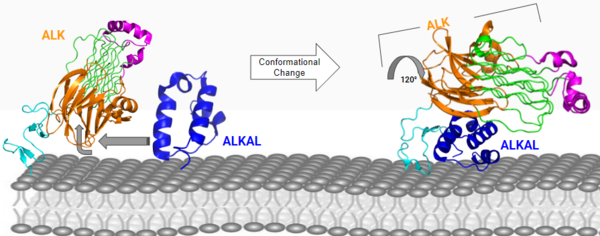
The value of the ALKr-ALKAL dimer is its rigidity and strength of its structure, being very important for the function of ALK. Each step of building from the ALK monomer to the dimer adds a level of strength in the structure, making it more likely to create the conformational change the kinase domain undergoes inside the cell across the membrane. This conformation into the dimer initiates a conformational change of the intracellular kinase domain of ALK. This causes an autophosphorylation of several tyrosine residues of this domain, which ultimately activates the kinase function of ALK and can now phosphorylate another protein or enzyme downstream in the signaling cascade. An example of a kinase that has a similar function to ALK are [https://proteopedia.org/wiki/index.php/Insulin_receptor#:~:text=The%20insulin%20receptor%20binds%20the,including%20skeletal%20muscle%20and%20adipose. insulin receptors] (IR). Their ligand (insulin) initiates a conformational change, which allows the kinase domain to be autophosphorylated, activating IR and allowing it to activate other enzymes or proteins down multiple possible signaling pathways via phosphorylation. | The value of the ALKr-ALKAL dimer is its rigidity and strength of its structure, being very important for the function of ALK. Each step of building from the ALK monomer to the dimer adds a level of strength in the structure, making it more likely to create the conformational change the kinase domain undergoes inside the cell across the membrane. This conformation into the dimer initiates a conformational change of the intracellular kinase domain of ALK. This causes an autophosphorylation of several tyrosine residues of this domain, which ultimately activates the kinase function of ALK and can now phosphorylate another protein or enzyme downstream in the signaling cascade. An example of a kinase that has a similar function to ALK are [https://proteopedia.org/wiki/index.php/Insulin_receptor#:~:text=The%20insulin%20receptor%20binds%20the,including%20skeletal%20muscle%20and%20adipose. insulin receptors] (IR). Their ligand (insulin) initiates a conformational change, which allows the kinase domain to be autophosphorylated, activating IR and allowing it to activate other enzymes or proteins down multiple possible signaling pathways via phosphorylation. | ||
== Disease == | == Disease == | ||
| - | Many <scene name='90/904317/Dimer_full_colored/9'>residual mutations</scene> have been identified that cause constitutive receptor activation. Enhanced receptor interaction or stabilization of active receptors also increase oncogenic potentials (Figure 3). One gain-of-function mutation found in [https://en.wikipedia.org/wiki/Adenocarcinoma_of_the_lung lung adenocarcinoma] is <scene name='90/904317/Dimer_full_colored/10'>His694</scene> that is mutated to arginine | + | Many <scene name='90/904317/Dimer_full_colored/9'>residual mutations</scene> have been identified that cause constitutive receptor activation. Enhanced receptor interaction or stabilization of active receptors also increase oncogenic potentials (Figure 3). One gain-of-function mutation found in [https://en.wikipedia.org/wiki/Adenocarcinoma_of_the_lung lung adenocarcinoma] is <scene name='90/904317/Dimer_full_colored/10'>His694</scene> that is mutated to arginine and leads to constitutive activation of ALK (Fig. 3b). What causes this gain-of-function is the increased length of arginine compared to histidine, which allows it to interact with the other ALKr-ALKAL complex via ionic bonding. This creates a stronger interaction between the two complexes, which increases the likelihood for ALK to be activated at any given time, which then also keeps the whole signal pathway activated downstream. Mutation of <scene name='90/904317/Dimer_full_colored/11'>Gly747</scene> to arginine could cause possible oncogenic potentials which are not specified yet (Fig. 3a), but this mutation would have a similar effect as His694Arg because it is an even bigger increase in length compared to the hydrogen side group of glycine. It is also a change from a nonpolar to a polar/charged side group, which will create ionic bonds with the other ALKr-ALKAL complex, making it an even bigger increase of interaction between the two complexes. The F856S and R753Q are two other mutations in ALK that are known to increase cytokine-dependent cell proliferation in certain cells. <ref>DOI:10.1038/s41586-021-03959-5</ref> |
[[Image:Screen Shot 2022-04-13 at 5.30.04 PM.png|800 px|left|thumb|Figure 3: Mutated residues on ALK that contribute to stabilization of the active state of ALK, leading to many types of cancers. A: Gly747Arg. B: His694Arg. [https://www.rcsb.org/structure/7N00 PDB: 7N00]]] | [[Image:Screen Shot 2022-04-13 at 5.30.04 PM.png|800 px|left|thumb|Figure 3: Mutated residues on ALK that contribute to stabilization of the active state of ALK, leading to many types of cancers. A: Gly747Arg. B: His694Arg. [https://www.rcsb.org/structure/7N00 PDB: 7N00]]] | ||
</StructureSection> | </StructureSection> | ||
Revision as of 02:12, 18 April 2022
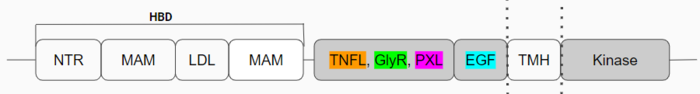
Anaplastic Lymphoma Kinase receptor
| |||||||||||
References
- ↑ Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ Jr, Shi X, Folta-Stogniew E, Gunel M, Alvarado D, Schlessinger J. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015 Jan 20;8(360):ra6. doi: 10.1126/scisignal.2005916. PMID:25605972 doi:http://dx.doi.org/10.1126/scisignal.2005916
- ↑ Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ De Munck S, Provost M, Kurikawa M, Omori I, Mukohyama J, Felix J, Bloch Y, Abdel-Wahab O, Bazan JF, Yoshimi A, Savvides SN. Structural basis of cytokine-mediated activation of ALK family receptors. Nature. 2021 Oct 13. pii: 10.1038/s41586-021-03959-5. doi:, 10.1038/s41586-021-03959-5. PMID:34646012 doi:http://dx.doi.org/10.1038/s41586-021-03959-5
Student Contributors
- Drew Peters
- Hillary Kulavic