We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1712
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
[[Image:Updated schematic.png|700 px|center|thumb|Figure 1: Anaplastic Lymphoma Kinase and its domains.]] | [[Image:Updated schematic.png|700 px|center|thumb|Figure 1: Anaplastic Lymphoma Kinase and its domains.]] | ||
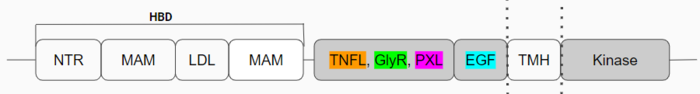
| - | ALKr is in its inactive state as a <scene name='90/904317/Glycinerichmonomer/12'>monomer</scene> and has many different domains (Figure 1) that are important to the formation of the <scene name='90/904317/Dimer_full_colored/12'>dimerized</scene> active state, leading to ALK's main function. The [https://en.wikipedia.org/wiki/Tumor_necrosis_factor tumor necrosis factor]-like domain (TNFL), glycine-rich domain (GlyR), polyglycine extension loop (PXL), and [https://en.wikipedia.org/wiki/Growth_factor growth factor]-like domain (EGF) are the main domains of ALKr | + | ALKr is in its inactive state as a <scene name='90/904317/Glycinerichmonomer/12'>monomer</scene> and has many different domains (Figure 1) that are important to the formation of the <scene name='90/904317/Dimer_full_colored/12'>dimerized</scene> active state, leading to ALK's main function. The [https://en.wikipedia.org/wiki/Tumor_necrosis_factor tumor necrosis factor]-like domain (TNFL), glycine-rich domain (GlyR), polyglycine extension loop (PXL), and [https://en.wikipedia.org/wiki/Growth_factor growth factor]-like domain (EGF) are the main domains of ALKr and the only domains whose structures have been fully discovered are in color (Figure 1). The <scene name='90/904317/Glycinerichmonomer/11'>EGF</scene> (cyan) is the domain that binds to the TMH (transmembrane region), connecting the extracellular portion of ALK to the intracellular kinase domain. <scene name='90/904317/Glycinerichmonomer/10'>TNFL</scene> (orange) has a [https://en.wikipedia.org/wiki/Beta-sandwich beta-sandwich] structure that provides important residues that act as the binding surface for the ligand. <scene name='90/904317/Glycinerichmonomer/13'>GlyR</scene> (green) contains 14 rare polyglycine helices that are hydrogen-bound to each other. The <scene name='90/904317/Glycinerichdomain/5'>hexagonal orientations</scene> of these rare helices create a rigid structure which allows it to function as a scaffold to anchor the ligand-binding site on the TNF-like domain while remaining bound to the ligand. The numerous hydrogen bonds create this <scene name='90/904317/Glycinerichdomain/3'>formation</scene> of the helices and the rigidity of the structure. The <scene name='90/904317/Glycinerichmonomer/9'>PXL</scene> (pink) connects two of these polyglycine helices, which plays a role in forming important interactions of the dimerized activated state of ALKr. |
Additional domains are present in ALK monomers (Figure 1), but their structures are not currently known. The [https://en.wikipedia.org/wiki/Heparin heparin] binding domains (HBDs), are at the N-terminal end of the monomer. Heparin is a likely activating ligand of ALK.<ref>DOI: 10.1126/scisignal.2005916</ref> The transmembrane domain (TMH) contains the membrane spanning portion of ALK that transmits extracellular ligand binding into an intracellular signal. The kinase domain is the intracellular portion of ALK that contains the Tyr residues which are auto-phosphorylated when ALK is activated, initiating a signaling cascade. <ref>DOI: 10.1038/s41586-021-04141-7</ref> | Additional domains are present in ALK monomers (Figure 1), but their structures are not currently known. The [https://en.wikipedia.org/wiki/Heparin heparin] binding domains (HBDs), are at the N-terminal end of the monomer. Heparin is a likely activating ligand of ALK.<ref>DOI: 10.1126/scisignal.2005916</ref> The transmembrane domain (TMH) contains the membrane spanning portion of ALK that transmits extracellular ligand binding into an intracellular signal. The kinase domain is the intracellular portion of ALK that contains the Tyr residues which are auto-phosphorylated when ALK is activated, initiating a signaling cascade. <ref>DOI: 10.1038/s41586-021-04141-7</ref> | ||
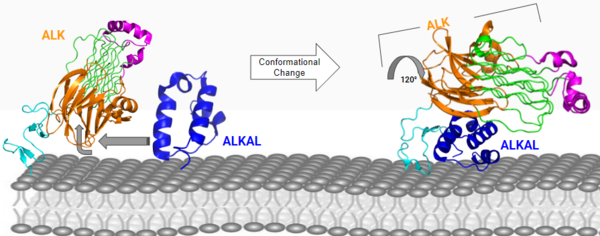
[[Image:Con chang 2D.png|600 px|center|thumb|Figure 2: ALK-ALKAL complex, showing the conformation change of ALK from the binding of ALKAL. [https://www.rcsb.org/structure/7N00 PDB: 7N00]]] | [[Image:Con chang 2D.png|600 px|center|thumb|Figure 2: ALK-ALKAL complex, showing the conformation change of ALK from the binding of ALKAL. [https://www.rcsb.org/structure/7N00 PDB: 7N00]]] | ||
Revision as of 21:46, 18 April 2022
Anaplastic Lymphoma Kinase receptor
| |||||||||||
References
- ↑ Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ Jr, Shi X, Folta-Stogniew E, Gunel M, Alvarado D, Schlessinger J. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015 Jan 20;8(360):ra6. doi: 10.1126/scisignal.2005916. PMID:25605972 doi:http://dx.doi.org/10.1126/scisignal.2005916
- ↑ Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ De Munck S, Provost M, Kurikawa M, Omori I, Mukohyama J, Felix J, Bloch Y, Abdel-Wahab O, Bazan JF, Yoshimi A, Savvides SN. Structural basis of cytokine-mediated activation of ALK family receptors. Nature. 2021 Oct 13. pii: 10.1038/s41586-021-03959-5. doi:, 10.1038/s41586-021-03959-5. PMID:34646012 doi:http://dx.doi.org/10.1038/s41586-021-03959-5
Student Contributors
- Drew Peters
- Hillary Kulavic