We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1701
From Proteopedia
(Difference between revisions)
| Line 42: | Line 42: | ||
==== Toggle Switch ==== | ==== Toggle Switch ==== | ||
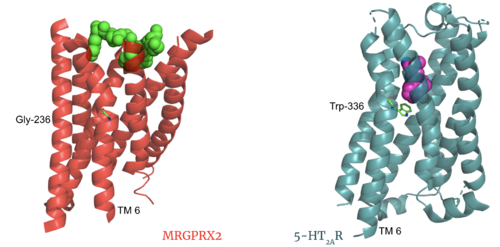
| - | Key toggle switch residues in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off". Toggle switches initiate the transmission of the molecular signal through the 7 TMD helices to the intracellular G protein. Trp-336 is the "iconic" toggle switch in class A GPCR’s <ref name="Trzaskowski">PMID: 22300046</ref>, and is a part of another motif, known as the '''CWxP motif'''. However in MRGPRX2, this tryptophan has been replaced with a <scene name='90/904305/Glycine_toggle_switch/ | + | Key toggle switch residues in the ligand binding pocket can act as molecular switches to turn the GPCR “on” or “off". Toggle switches initiate the transmission of the molecular signal through the 7 TMD helices to the intracellular G protein. Trp-336 is the "iconic" toggle switch in class A GPCR’s <ref name="Trzaskowski">PMID: 22300046</ref>, and is a part of another motif, known as the '''CWxP motif'''. However in MRGPRX2, this tryptophan has been replaced with a <scene name='90/904305/Glycine_toggle_switch/8'>glycine</scene> <ref name="Cao">PMID: 34789874</ref> <ref name="Yang">PMID: 34789875</ref>. This shift leads to a significant modification to the receptor structure. By replacing the large tryptophan residue with a small glycine, the membrane helices, especially helix 7 on which the toggle switch is found, pack more tightly. The ligands that interact with MRGPRX2 are able to bind much <scene name='90/904305/Glycine_toggle_switch_and_cor/4'>closer to the surface</scene> of the receptor, as opposed to deeper within the helices ('''Figure 3'''). This shallower binding pocket expands the types of ligands that are able to interact with X2 and therefore what types of molecules can activate the Human Itch GPCR. More details about what kinds of ligands bind to this receptor are discussed later. |
<scene name='90/904306/Alignment_toggle/1'>Toggle Switch Alignment</scene> | <scene name='90/904306/Alignment_toggle/1'>Toggle Switch Alignment</scene> | ||
| Line 56: | Line 56: | ||
</jmol> | </jmol> | ||
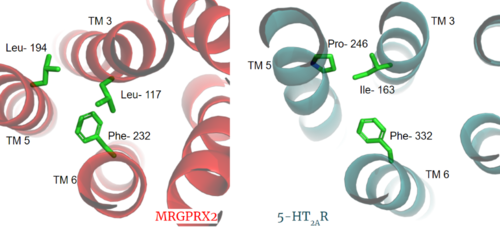
[[Image:PIF_resized.png|500px|right|thumb|'''Figure 3.''' Conserved PIF motif in 5HT2AR (teal) compared to the LLF motif found in MRGPRX2 (red). Transmembrane helices and residues are numbered and labeled to show how this structural change shifts the orientation of the helices.]] | [[Image:PIF_resized.png|500px|right|thumb|'''Figure 3.''' Conserved PIF motif in 5HT2AR (teal) compared to the LLF motif found in MRGPRX2 (red). Transmembrane helices and residues are numbered and labeled to show how this structural change shifts the orientation of the helices.]] | ||
| - | Another motif found in most, but not all, A family GPCR’s is the PIF motif. The PIF residues (<scene name='90/904305/Llf_motif/ | + | Another motif found in most, but not all, A family GPCR’s is the PIF motif. The PIF residues (<scene name='90/904305/Llf_motif/5'>Leu-117, Leu-194, and Phe-232</scene>) are found on transmembrane helices 5, 3, and 6, respectively. In MRGPRX2, the PIF motif is changed to LLF residues. '''Figure 3''' shows the conserved PIF motif on 5HT2AR, <scene name='90/904306/Alignment_llf/1'>compared</scene> to the LLF motif on MRGPRX2. This change to LLF shifts helix 6 towards helix 3, and contributes to the tighter packing of helices <ref name="Cao">PMID: 34789874</ref> <ref name="Yang">PMID: 34789875</ref> and therefore a more surface-level ligand binding site. |
| Line 73: | Line 73: | ||
==== Disulfide Bonds ==== | ==== Disulfide Bonds ==== | ||
[[Image:Screen Shot 2022-03-27 at 5.45.52 PM.png|300px|right|thumb|'''Figure 5.''' Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.]] | [[Image:Screen Shot 2022-03-27 at 5.45.52 PM.png|300px|right|thumb|'''Figure 5.''' Overlay of the 5HT2AR and MRGPRX2 TMP for comparison of disulfide bond location.]] | ||
| - | In a large majority of class A GPCRs, there is a conserved disulfide bond between extracellular loop 2 (ECL2) and transmembrane helix 3 (TM3). This bond has been proposed to have a role in structural stability, expression, and function of GPCRs<ref name="Naranjo">PMID: 25445670</ref>. The MRGPRX2 disulfide bond is between <scene name='90/904305/Disulfide_bond/ | + | In a large majority of class A GPCRs, there is a conserved disulfide bond between extracellular loop 2 (ECL2) and transmembrane helix 3 (TM3). This bond has been proposed to have a role in structural stability, expression, and function of GPCRs<ref name="Naranjo">PMID: 25445670</ref>. The MRGPRX2 disulfide bond is between <scene name='90/904305/Disulfide_bond/2'>Cys-168 and Cys-180</scene> on TM helices 5 and 4, respectively. For example, the <scene name='90/904306/5ht2a_disulfide/2'>serotonin GPCR</scene> shows this disulfide bond between the ECL2 and TM3. Although this bond is in a different location than other class A GPCRs, there is evidence to suggest its location is essential for the signaling of the X2 receptor as the ECL2 instead located at the top of TM4 and TM5 allowing for the large, extracellular binding pocket observed in X2<ref name="Cao">PMID: 34789874</ref>. |
<scene name='90/904306/Alignment_disulfide/1'>Disulfide Bond Alignment</scene> | <scene name='90/904306/Alignment_disulfide/1'>Disulfide Bond Alignment</scene> | ||
Revision as of 23:41, 18 April 2022
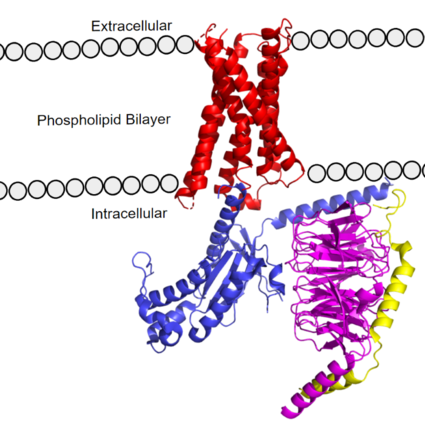
MRGPRX2 Human Itch G-Protein Coupled Receptor (GPCR)
| |||||||||||
References
- ↑ Tuteja N. Signaling through G protein coupled receptors. Plant Signal Behav. 2009 Oct;4(10):942-7. doi: 10.4161/psb.4.10.9530. Epub 2009, Oct 14. PMID:19826234 doi:http://dx.doi.org/10.4161/psb.4.10.9530
- ↑ Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub, 2017 Oct 27. PMID:29075003 doi:http://dx.doi.org/10.1038/nrd.2017.178
- ↑ 3.0 3.1 3.2 3.3 3.4 Porebski G, Kwiecien K, Pawica M, Kwitniewski M. Mas-Related G Protein-Coupled Receptor-X2 (MRGPRX2) in Drug Hypersensitivity Reactions. Front Immunol. 2018 Dec 20;9:3027. doi: 10.3389/fimmu.2018.03027. eCollection, 2018. PMID:30619367 doi:http://dx.doi.org/10.3389/fimmu.2018.03027
- ↑ 4.0 4.1 4.2 4.3 Dondalska A, Ronnberg E, Ma H, Palsson SA, Magnusdottir E, Gao T, Adam L, Lerner EA, Nilsson G, Lagerstrom M, Spetz AL. Amelioration of Compound 48/80-Mediated Itch and LL-37-Induced Inflammation by a Single-Stranded Oligonucleotide. Front Immunol. 2020 Sep 30;11:559589. doi: 10.3389/fimmu.2020.559589. eCollection, 2020. PMID:33101278 doi:http://dx.doi.org/10.3389/fimmu.2020.559589
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 McNeil BD, Pundir P, Meeker S, Han L, Undem BJ, Kulka M, Dong X. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 2015 Mar 12;519(7542):237-41. doi: 10.1038/nature14022. Epub 2014 Dec 17. PMID:25517090 doi:http://dx.doi.org/10.1038/nature14022
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, Slocum ST, Krumm BE, Lansu K, McCorvy JD, Kroeze WK, English JG, DiBerto JF, Olsen RHJ, Huang XP, Zhang S, Liu Y, Kim K, Karpiak J, Jan LY, Abraham SN, Jin J, Shoichet BK, Fay JF, Roth BL. Structure, function and pharmacology of human itch GPCRs. Nature. 2021 Dec;600(7887):170-175. doi: 10.1038/s41586-021-04126-6. Epub 2021, Nov 17. PMID:34789874 doi:http://dx.doi.org/10.1038/s41586-021-04126-6
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 7.6 7.7 7.8 7.9 Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP. Structure, function and pharmacology of human itch receptor complexes. Nature. 2021 Dec;600(7887):164-169. doi: 10.1038/s41586-021-04077-y. Epub 2021, Nov 17. PMID:34789875 doi:http://dx.doi.org/10.1038/s41586-021-04077-y
- ↑ Kamato D, Thach L, Bernard R, Chan V, Zheng W, Kaur H, Brimble M, Osman N, Little PJ. Structure, Function, Pharmacology, and Therapeutic Potential of the G Protein, Galpha/q,11. Front Cardiovasc Med. 2015 Mar 24;2:14. doi: 10.3389/fcvm.2015.00014. eCollection, 2015. PMID:26664886 doi:http://dx.doi.org/10.3389/fcvm.2015.00014
- ↑ Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090-109. doi: 10.2174/092986712799320556. PMID:22300046 doi:http://dx.doi.org/10.2174/092986712799320556
- ↑ 10.0 10.1 Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014 May;39(5):233-44. doi: 10.1016/j.tibs.2014.03.002. Epub , 2014 Apr 21. PMID:24767681 doi:http://dx.doi.org/10.1016/j.tibs.2014.03.002
- ↑ Rovati GE, Capra V, Neubig RR. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol. 2007 Apr;71(4):959-64. doi: 10.1124/mol.106.029470. Epub 2006 Dec , 27. PMID:17192495 doi:http://dx.doi.org/10.1124/mol.106.029470
- ↑ Naranjo AN, Chevalier A, Cousins GD, Ayettey E, McCusker EC, Wenk C, Robinson AS. Conserved disulfide bond is not essential for the adenosine A2A receptor: Extracellular cysteines influence receptor distribution within the cell and ligand-binding recognition. Biochim Biophys Acta. 2015 Feb;1848(2):603-14. doi: 10.1016/j.bbamem.2014.11.010., Epub 2014 Nov 16. PMID:25445670 doi:http://dx.doi.org/10.1016/j.bbamem.2014.11.010
- ↑ Olivella M, Caltabiano G, Cordomi A. The role of Cysteine 6.47 in class A GPCRs. BMC Struct Biol. 2013 Mar 15;13:3. doi: 10.1186/1472-6807-13-3. PMID:23497259 doi:http://dx.doi.org/10.1186/1472-6807-13-3