We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| Line 33: | Line 33: | ||
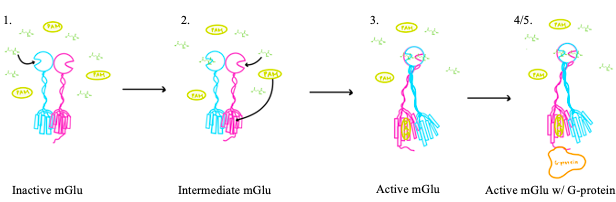
[[Image:Glutamate.png|300 px|right|thumb|Figure 1. Structure of glutamate under physiological conditions (pH 7.4).]] To activate the mGlu transformation, glutamate acts as the protein's main agonist. Glutamate is an acidic, polar amino acid (Figure 1). This agonist binds to the extracellular portion of the glutamate receptor causing the transmembrane spanning region of the homodimer to change conformationally. This change allows for the mGlu to bind to a G-protein which initiates a signaling cascade within the cell that can ultimately lead to the modification of other proteins and a difference in the synapse’s excitability<ref name="Seven">PMID:34194039</ref>. In mGlu, the binding affinity of glutamate is also controlled by the binding of either a positive (PAM) or negative (NAM) allosteric modulator to a binding pocket within the seven TMD<ref name="Lin">PMID:34135510</ref>. | [[Image:Glutamate.png|300 px|right|thumb|Figure 1. Structure of glutamate under physiological conditions (pH 7.4).]] To activate the mGlu transformation, glutamate acts as the protein's main agonist. Glutamate is an acidic, polar amino acid (Figure 1). This agonist binds to the extracellular portion of the glutamate receptor causing the transmembrane spanning region of the homodimer to change conformationally. This change allows for the mGlu to bind to a G-protein which initiates a signaling cascade within the cell that can ultimately lead to the modification of other proteins and a difference in the synapse’s excitability<ref name="Seven">PMID:34194039</ref>. In mGlu, the binding affinity of glutamate is also controlled by the binding of either a positive (PAM) or negative (NAM) allosteric modulator to a binding pocket within the seven TMD<ref name="Lin">PMID:34135510</ref>. | ||
== Structural Highlights == | == Structural Highlights == | ||
| - | mGlu receptors are dimeric proteins consisting of an <scene name='90/904320/Inactive_mglu2/ | + | mGlu receptors are dimeric proteins consisting of an <scene name='90/904320/Inactive_mglu2/10'>alpha and beta chain</scene>. While a heterodimer of different mGlu subtypes can form, only homodimeric receptors can become active<ref name="Seven">PMID:34194039</ref>. Both the alpha and beta chains are comprised of <scene name='90/904320/Mglu2_domains/7'>3 domains</scene>: the venus fly trap (VFT), cysteine rich domain (CRD), and the transmembrane domain (TMD). |
[[Image: Active vs inactive vft.png|250px|left|thumb|Figure 2. Cysteine 121 positioning in the inactive (left) vs the active (right) mGlu conformation.]] | [[Image: Active vs inactive vft.png|250px|left|thumb|Figure 2. Cysteine 121 positioning in the inactive (left) vs the active (right) mGlu conformation.]] | ||
Revision as of 01:36, 19 April 2022
Metabotropic Glutamate Receptor
| |||||||||||
3D Structures
7epa, mGlu Inactive
7mtr, mGlu Active
7mts, mGlu Active G Protein Bound
References
- ↑ Katritch V, Cherezov V, Stevens RC. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531-56. doi:, 10.1146/annurev-pharmtox-032112-135923. Epub 2012 Nov 8. PMID:23140243 doi:http://dx.doi.org/10.1146/annurev-pharmtox-032112-135923
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295-322. doi:, 10.1146/annurev.pharmtox.011008.145533. PMID:20055706 doi:http://dx.doi.org/10.1146/annurev.pharmtox.011008.145533
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 Seven AB, Barros-Alvarez X, de Lapeyriere M, Papasergi-Scott MM, Robertson MJ, Zhang C, Nwokonko RM, Gao Y, Meyerowitz JG, Rocher JP, Schelshorn D, Kobilka BK, Mathiesen JM, Skiniotis G. G-protein activation by a metabotropic glutamate receptor. Nature. 2021 Jun 30. pii: 10.1038/s41586-021-03680-3. doi:, 10.1038/s41586-021-03680-3. PMID:34194039 doi:http://dx.doi.org/10.1038/s41586-021-03680-3
- ↑ 4.0 4.1 4.2 4.3 Lin S, Han S, Cai X, Tan Q, Zhou K, Wang D, Wang X, Du J, Yi C, Chu X, Dai A, Zhou Y, Chen Y, Zhou Y, Liu H, Liu J, Yang D, Wang MW, Zhao Q, Wu B. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021 Jun;594(7864):583-588. doi: 10.1038/s41586-021-03495-2. Epub 2021, Jun 16. PMID:34135510 doi:http://dx.doi.org/10.1038/s41586-021-03495-2
- ↑ 5.0 5.1 5.2 5.3 5.4 Crupi R, Impellizzeri D, Cuzzocrea S. Role of Metabotropic Glutamate Receptors in Neurological Disorders. Front Mol Neurosci. 2019 Feb 8;12:20. doi: 10.3389/fnmol.2019.00020. eCollection , 2019. PMID:30800054 doi:http://dx.doi.org/10.3389/fnmol.2019.00020
- ↑ Bordi F, Ugolini A. Group I metabotropic glutamate receptors: implications for brain diseases. Prog Neurobiol. 1999 Sep;59(1):55-79. doi: 10.1016/s0301-0082(98)00095-1. PMID:10416961 doi:http://dx.doi.org/10.1016/s0301-0082(98)00095-1
- ↑ Conn PJ, Lindsley CW, Jones CK. Activation of metabotropic glutamate receptors as a novel approach for the treatment of schizophrenia. Trends Pharmacol Sci. 2009 Jan;30(1):25-31. doi: 10.1016/j.tips.2008.10.006. Epub, 2008 Dec 6. PMID:19058862 doi:http://dx.doi.org/10.1016/j.tips.2008.10.006
Student Contributors
- Courtney Vennekotter
- Cade Chezem