This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| Line 41: | Line 41: | ||
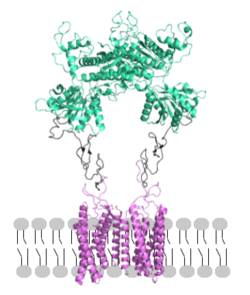
<scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the protomer that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between cysteines. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate to the TMD. The change resulting from the binding of glutamate in the VFT brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I <ref name="Seven">PMID:34194039</ref>. | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the protomer that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between cysteines. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate to the TMD. The change resulting from the binding of glutamate in the VFT brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I <ref name="Seven">PMID:34194039</ref>. | ||
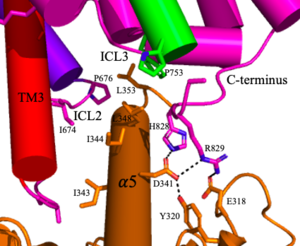
| - | <scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the hydrophobicity of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers. Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers <ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). | + | <scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the hydrophobicity of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers (Figure 3A). Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers (Figure3 B)<ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). |
[[Image:Screen Shot 2022-04-19 at 2.52.21 AM.png|500px|center|thumb|Figure 3. A) The inactive transmembrane helices conformation. B) The active transmembrane helices conformation.]] | [[Image:Screen Shot 2022-04-19 at 2.52.21 AM.png|500px|center|thumb|Figure 3. A) The inactive transmembrane helices conformation. B) The active transmembrane helices conformation.]] | ||
| Line 51: | Line 51: | ||
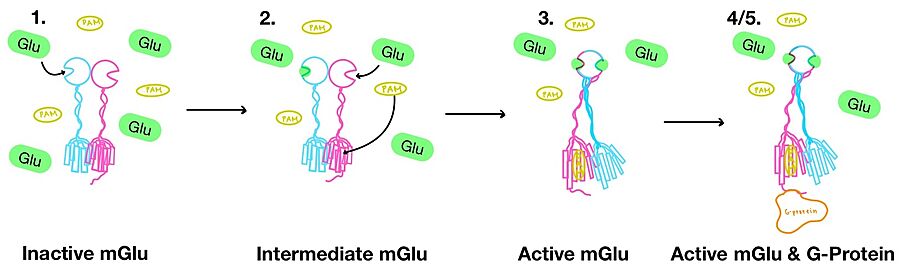
[[Image:Overview_mGlu_2.jpg|900 px|center|thumb|Figure 4. Illustration of mGlu's conformational change process.]] | [[Image:Overview_mGlu_2.jpg|900 px|center|thumb|Figure 4. Illustration of mGlu's conformational change process.]] | ||
| - | '''3.''' A second glutamate then binds to the other <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> of the VFT. Mediated by L639, F643, N735, W773, and F776, a <scene name='90/904320/Pam/8'>positive allosteric modulator</scene> (PAM) also binds within the seven TMD helices of the alpha chain <ref name="Seven">PMID:34194039</ref>. This closed conformation of the VFT now has an inter-lobe angle of 25° is considered to be in the <scene name='90/904320/Active_mglu/10'>active conformation</scene><ref name="Seven">PMID:34194039</ref>. The binding of these ligands allows the CRDs to compact and come together. This transformation causes the TMD to form a separate, active asymmetric conformation with a <scene name='90/904320/Active_helices/14'>TM6-TM6 interface</scene> between the chains<ref name="Seven">PMID:34194039</ref>. | + | '''3.''' A second glutamate then binds to the other <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> of the VFT. Mediated by L639, F643, N735, W773, and F776, a <scene name='90/904320/Pam/8'>positive allosteric modulator</scene> (PAM) also binds within the seven TMD helices of the alpha chain <ref name="Seven">PMID:34194039</ref>. This closed conformation of the VFT now has an inter-lobe angle of 25° is considered to be in the <scene name='90/904320/Active_mglu/10'>active conformation</scene><ref name="Seven">PMID:34194039</ref>. The binding of these ligands allows the CRDs to compact and come together. This transformation causes the TMD to form a separate, active asymmetric conformation with a <scene name='90/904320/Active_helices/14'>TM6-TM6 interface</scene> between the chains (Figure 3)<ref name="Seven">PMID:34194039</ref>. |
[[Image:Screen Shot 2022-04-18 at 10.20.26 PM.png|300 px|right|thumb|Figure 5. The interaction between an active mGlu (magenta/lime/purple/crimson) and a G-protein (orange). Hydrogen bonds are shown through black dashes]] | [[Image:Screen Shot 2022-04-18 at 10.20.26 PM.png|300 px|right|thumb|Figure 5. The interaction between an active mGlu (magenta/lime/purple/crimson) and a G-protein (orange). Hydrogen bonds are shown through black dashes]] | ||
Revision as of 07:12, 19 April 2022
Metabotropic Glutamate Receptor
| |||||||||||
Student Contributors
- Courtney Vennekotter
- Cade Chezem