We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1706
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
==Introduction== | ==Introduction== | ||
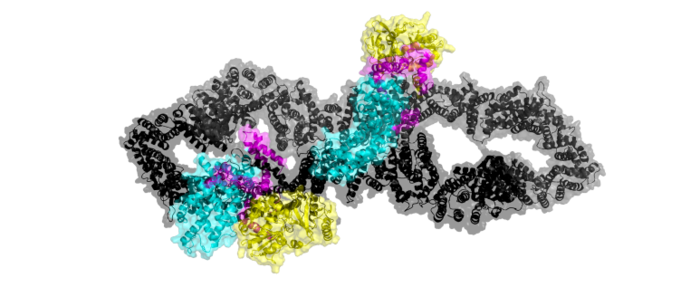
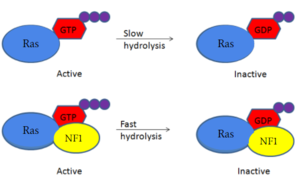
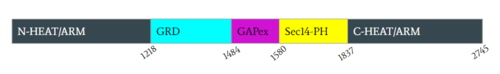
| - | [https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin. Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MAPK/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Decreased activity of neurofibromin due to mutation can lead to tumor growth along nerves. As it is ubiquitous expression, NF1 misregulation can cause systemic tumor growth. Neurofibromin is localized to the [https://en.wikipedia.org/wiki/Cytosol cytosol] but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to inactivate [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was determined by high-resolution single particle [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryo-EM]. These structures illustrated the domain architecture and conformational changes in neurofibromin, controlling Ras binding and inactivation. <ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | [https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref> Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MAPK/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Decreased activity of neurofibromin due to mutation can lead to tumor growth along nerves. As it is ubiquitous expression, NF1 misregulation can cause systemic tumor growth. Neurofibromin is localized to the [https://en.wikipedia.org/wiki/Cytosol cytosol] but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to inactivate [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was determined by high-resolution single particle [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryo-EM]. These structures illustrated the domain architecture and conformational changes in neurofibromin, controlling Ras binding and inactivation. <ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
==Function== | ==Function== | ||
| Line 33: | Line 33: | ||
====Closed conformation==== | ====Closed conformation==== | ||
| - | Ras is unable to bind to the GRD binding site when both of the neurofibromin protomers are in the <scene name='90/904311/ | + | Ras is unable to bind to the GRD binding site when both of the neurofibromin protomers are in the <scene name='90/904311/Closed_conformation/8'>closed conformation</scene>. In the closed conformation, one protomer has its domains shifted by a 130° rotation of three separate conformational change linkers. That rotation places Arg1276 in an orientation where binding of Y32 of Ras is sterically hindered by E31. (<scene name='90/904311/Closed_arg/6'>Arg1276, Y32, and E31 interaction</scene> |
<jmol> | <jmol> | ||
<jmolButton> | <jmolButton> | ||
| Line 81: | Line 81: | ||
==Clinical Relevance== | ==Clinical Relevance== | ||
| - | [https://en.wikipedia.org/wiki/Germline_mutation Germline mutations] are common in NF1 and often cause genetic tumor syndrome through misregulation of the Ras signaling pathway. [https://en.wikipedia.org/wiki/Somatic_mutation Somatic mutations] among NF1 are also extremely common. In germline mutations and some somatic mutations of NF1, tumors develop along the deep epidermis layer of the skin. Understanding how mutations affect the structure and function of neurofibromin will allow for advancements in treatment. | + | [https://en.wikipedia.org/wiki/Germline_mutation Germline mutations] are common in NF1 and often cause genetic tumor syndrome through misregulation of the Ras signaling pathway. <ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref> [https://en.wikipedia.org/wiki/Somatic_mutation Somatic mutations] among NF1 are also extremely common. In germline mutations and some somatic mutations of NF1, tumors develop along the deep epidermis layer of the skin. Understanding how mutations affect the structure and function of neurofibromin will allow for advancements in treatment. |
<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | <ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | ||
Revision as of 12:11, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Neurofibromin 1
| |||||||||||