Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| Line 39: | Line 39: | ||
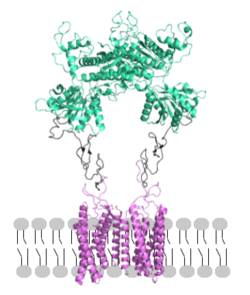
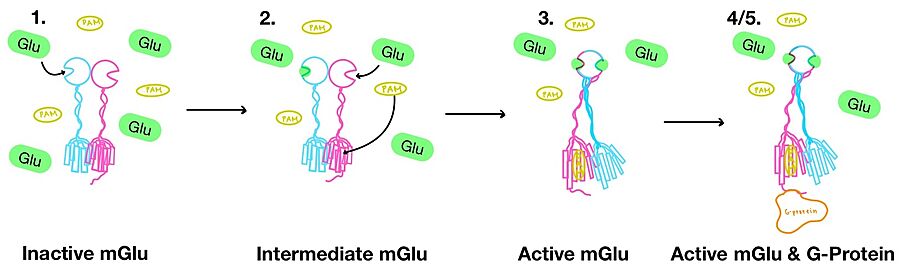
<scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains. This <scene name='90/904320/Inactive_mglu/13'>disulfide bond</scene> is shifted down in the open conformation and undergoes a <scene name='90/904320/Cys_active/3'>disulfide bond upward movement</scene> upon glutamate binding which stabilizes the <scene name='90/904319/Vft/2'>active site</scene>. Glutamate binds within the <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> via [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding], specifically hydrogen bonding, with R57, S143, S145, T168, and K377 of the VFT. This binding initiates a closed VFT conformation. | <scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains. This <scene name='90/904320/Inactive_mglu/13'>disulfide bond</scene> is shifted down in the open conformation and undergoes a <scene name='90/904320/Cys_active/3'>disulfide bond upward movement</scene> upon glutamate binding which stabilizes the <scene name='90/904319/Vft/2'>active site</scene>. Glutamate binds within the <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> via [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding], specifically hydrogen bonding, with R57, S143, S145, T168, and K377 of the VFT. This binding initiates a closed VFT conformation. | ||
| - | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the protomer that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between [https://en.wikipedia.org/wiki/Cysteine cysteines]. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. | + | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the [https://en.wikipedia.org/wiki/Protomer protomer] that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between [https://en.wikipedia.org/wiki/Cysteine cysteines]. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. |
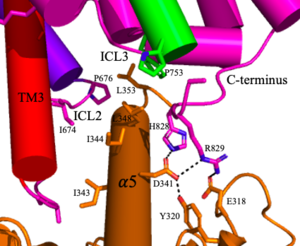
<scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane<ref name="Niswender">PMID:20055706</ref>. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the hydrophobicity of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers (Figure 3A). Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered<ref name="Seven">PMID:34194039</ref>. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers (Figure 3B)<ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G-protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). | <scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane<ref name="Niswender">PMID:20055706</ref>. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the hydrophobicity of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers (Figure 3A). Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered<ref name="Seven">PMID:34194039</ref>. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers (Figure 3B)<ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G-protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). | ||
Revision as of 14:41, 19 April 2022
Metabotropic Glutamate Receptor
| |||||||||||
Student Contributors
- Courtney Vennekotter
- Cade Chezem