We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1722
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
==== ''Toggle Switch'' ==== | ==== ''Toggle Switch'' ==== | ||
| - | In β2AR, and other Class A GPCRs, a “toggle switch” of <scene name='90/904327/B2artoggleswitchyes/7'>Trp-286</scene> | + | In β2AR, and other Class A GPCRs, a “toggle switch” of <scene name='90/904327/B2artoggleswitchyes/7'>Trp-286</scene> limits the proximity of the TM helices as tryptophan sterically occludes tight interaction. This results in a deep binding pocket for ligand binding. In contrast, in MRGPRX2 Trp-286 is replaced by <scene name='90/904327/Toggle_switch_gly_pt_2/2'>Gly-236</scene> <ref name="Cao"/> <ref name="Yang"/>. Glycine is a much smaller amino acid and thus allows the helices to close the base of the binding pocket. This causes MRGPRX2 to have a much shallower binding site and allows more promiscuous ligand binding. This can be seen in a shorter distance from the toggle switch to the ligand in β2AR (0.573 nm) compared to MRGPRX2 (1.389 nm). |
==== ''Disulfide bonds'' ==== | ==== ''Disulfide bonds'' ==== | ||
| - | In common Class A GPCRs the disulfide bond associated with the initiation of signal transduction is located on the extracellular domain of the 7 transmembrane helices | + | In common Class A GPCRs the disulfide bond associated with the initiation of signal transduction is located on the extracellular domain of the 7 transmembrane helices <ref name="Zhang">PMID: 26467290</ref>. The close proximity of the disulfide bond to the ligand binding site, allows the receptor to know that it is being bound and initiate the signal through the rest of the receptor. The <scene name='90/904328/B2ardisulfidebond1_pt_3/1'>disulfide bond of β2AR</scene>, a well-studied Class A GPCR, occurs between transmembrane three (TM3) Cys-106 and an extracellular loop (EL) Cys-191. This loop crosses through the middle of the extracellular domain, creating a barrier for bulkier substrates. The <scene name='90/904327/7tm_domain_pt_6/1'>disulfide bond of MRGPRX2</scene> is located between (TM4) Cys-168 and (TM5) Cys-180. This is a TM to TM disulfide bond as compared to a TM to EL disulfide bond seen in typical Class A GPCRs. This lack of interaction with the extracellular loop seen in MRGPRX2 causes the extracellular loop to flip on top of the TM4 and TM5 resulting in an open space for larger substrates to be able to interact with the receptor. |
==== ''PIF Motif'' ==== | ==== ''PIF Motif'' ==== | ||
| - | MRGPRX2 also differs from typical Class A GPCRs based on substitutions in the PIF/connector motif, which acts as a microswitch. PIF plays a role in connecting the binding pocket to conformational rearrangements required for receptor activation<ref name="Schonegge">Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x</ref>. The PIF motif is located towards the base of the TM domains. In a Class A GPCR, like <scene name='90/904327/B2arpif_pt_2/1'>β2AR, the PIF motif</scene> consists of Phe-211, Ile-121, and Phe-282 on TM domains 5, 3, and 6, respectively. However, for <scene name='90/904327/Mxpifnolabel_pt_3/1'>MRGPRX2, this PIF motif</scene> is replaced with Leu-194 on TM5, Leu-117 on TM3, and Phe-232 on TM6. | + | MRGPRX2 also differs from typical Class A GPCRs based on substitutions in the PIF/connector motif, which acts as a microswitch. PIF plays a role in connecting the binding pocket to conformational rearrangements required for receptor activation<ref name="Schonegge">Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x</ref>. The PIF motif is located towards the base of the TM domains. In a Class A GPCR, like <scene name='90/904327/B2arpif_pt_2/1'>β2AR, the PIF motif</scene> consists of Phe-211, Ile-121, and Phe-282 on TM domains 5, 3, and 6, respectively. However, for <scene name='90/904327/Mxpifnolabel_pt_3/1'>MRGPRX2, this PIF motif</scene> is replaced with Leu-194 on TM5, Leu-117 on TM3, and Phe-232 on TM6. These modifications result in tighter packing around the base of the receptor, indicating slightly different interactions with the intracellular [https://en.wikipedia.org/wiki/G_protein G-proteins]. |
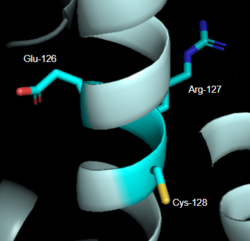
| - | [[Image:LabeledDRY.PNG|250px|right|thumb|'''Figure 2''': DRY Motif of MRGPRX2 | + | [[Image:LabeledDRY.PNG|250px|right|thumb|'''Figure 2''': DRY Motif of MRGPRX2 <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>]].(PDB entry:[https://www.rcsb.org/structure/7S8L 7S8L] |
==== ''DRY Motif'' ==== | ==== ''DRY Motif'' ==== | ||
| - | The DRY motif is a proton microswitch that is located near the G-protein binding site C-terminal | + | The DRY motif is a proton microswitch that is located near the G-protein binding site C-terminal on TM3 <ref name="Schonegge"/>. It acts as lock when the GPCR is not being activated, preventing unnecessary activation of the G-proteins <ref name="Zhou"/>. This motif is conserved in typical Class A GPCRS however, in MRGPRX2 it is only partially conserved. The arginine is conserved, while the aspartate is replaced by a glutamate and the tyrosine is replaced by a cysteine <ref name="Yang"/> <ref name="Sandoval">Sandoval, A., et al. "The Molecular Switching Mechanism at the Conserved D(E)RY Motif in Class-A GPCRs." Biophysical journal, 111(1), 79-89. https://doi.org/10.1016/j.bpj.2016.06.004 </ref> (Figure 2). The replacement of tyrosine for cysteine results in the helices coming closer together, creating a shallower binding pocket<ref name="Zhou"/>. |
==== ''Sodium Binding'' ==== | ==== ''Sodium Binding'' ==== | ||
| - | The sodium site motif facilitates the conformational change of GPCR upon activation | + | The sodium site motif facilitates the conformational change of GPCR upon activation<ref name="Katritch">PMID: 24767681</ref>. A sodium molecule sits in the middle of the TM7 helices where it is stabilized by conserved residues aspartate (TM2), serine (TM2), and three water molecules. The sodium is able to make a salt bridge with the aspartate at this position. The sodium acts similar to a ball joint in which it allows for the TM helices be spread apart and induce larger conformational change upon binding. In MRGPRX2, this motif is only partially conserved. The aspartate (TM2) is conserved while the serine is replaced by a glycine <ref name="Yang"/> .This creates a less favorable environment for the stabilization of sodium due to serine being polar while glycine is nonpolar. Currently, in crystallization structures no sodium has been seen at this site. Thus making it inconclusive on whether it plays a role in the conformational change to activate G-proteins upon binding to the receptor <ref name="Yang"/> . |
== MRGPRX2 Signaling Pathway == | == MRGPRX2 Signaling Pathway == | ||
| Line 47: | Line 47: | ||
=== 1. Binding Pocket === | === 1. Binding Pocket === | ||
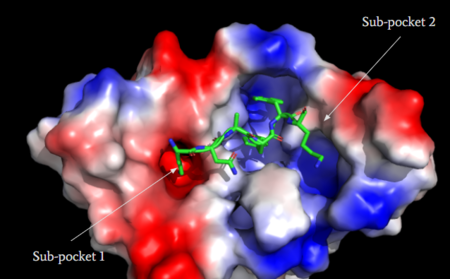
| - | MRGPRX2 consists of two binding pockets (seen in Figure 3). Sub-pocket 1 consists of acidic catalytic residues Asp-184 and Glu-164 that interact with substrates by making ion pairs. There are also some hydrophobic aromatic residues, Phe-170, Trp-243, and Phe-244, towards the top of the binding pocket | + | MRGPRX2 consists of two binding pockets (seen in Figure 3). Sub-pocket 1 consists of acidic catalytic residues Asp-184 and Glu-164 that interact with substrates by making ion pairs. There are also some hydrophobic aromatic residues, Phe-170, Trp-243, and Phe-244, towards the top of the binding pocket <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>. These residues provide stabilization with ligands through stacking. Lastly, this pocket is in close proximity with the commonly conserved disulfide bond (formed by Cys-168 and Cys-180) seen in most Class A GPCRs. The second binding pocket forms hydrophobic interactions with larger substrates (seen in Figure 2) but is generally less studied <ref name="Cao"/>. |
[[Image:Electro.PNG|450px|center|thumb|'''Figure 3''': Binding pocket of MRGPRX2 with cortistatin-14. Two different binding pockets are present in MRGPRX2 and cortistatin-14 interacts with both of them. <ref name="Cao"/>]] | [[Image:Electro.PNG|450px|center|thumb|'''Figure 3''': Binding pocket of MRGPRX2 with cortistatin-14. Two different binding pockets are present in MRGPRX2 and cortistatin-14 interacts with both of them. <ref name="Cao"/>]] | ||
==== Agonists ==== | ==== Agonists ==== | ||
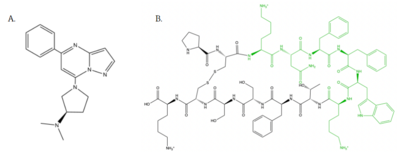
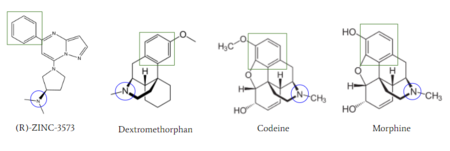
| - | [[Image:Agoniststogether.PNG |400px|right|thumb|'''Figure 4''': Structure of MRGPRX2 Agonists. (A) Structure of R-Zinc 3573. A cationic ligand selected for binding to MRGPRX2 | + | [[Image:Agoniststogether.PNG |400px|right|thumb|'''Figure 4''': Structure of MRGPRX2 Agonists. (A) Structure of R-Zinc 3573. A cationic ligand selected for binding to MRGPRX2 <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>.(B) Structure of Cortistatin-14 with resolved amino acids highlighted in green. These ligands were used as a probes for MRGPRX2 function and stabilization for structure determination <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>.]] |
===== Small Molecule ===== | ===== Small Molecule ===== | ||
| Line 66: | Line 66: | ||
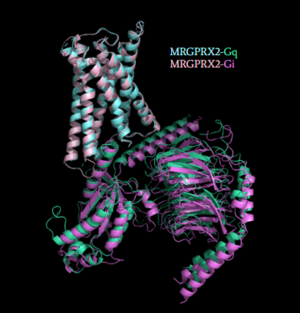
[[Image:Gq_and_Gi_snip.PNG|300px|right|thumb|'''Figure 5''': Comparison of the conformational change for MRGPRX2 (light blue) coupled with Gq (teal) and MRGPRX2 (pink) coupled with Gi (purple).<ref name="Cao"/> PDB entry [https://www.rcsb.org/structure/7S8N 7S8N] and [https://www.rcsb.org/structure/7S8O 7S8O]]] | [[Image:Gq_and_Gi_snip.PNG|300px|right|thumb|'''Figure 5''': Comparison of the conformational change for MRGPRX2 (light blue) coupled with Gq (teal) and MRGPRX2 (pink) coupled with Gi (purple).<ref name="Cao"/> PDB entry [https://www.rcsb.org/structure/7S8N 7S8N] and [https://www.rcsb.org/structure/7S8O 7S8O]]] | ||
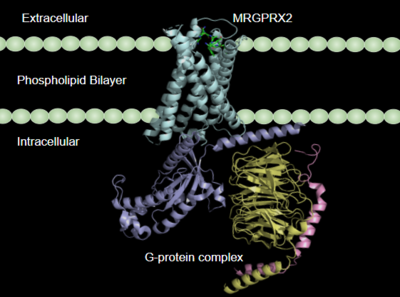
| - | Once the ligand is bound, MRGPRX2 undergoes a conformational change that is transmitted through to the [https://en.wikipedia.org/wiki/G_protein#:~:text=G%20proteins%2C%20also%20known%20as,a%20cell%20to%20its%20interior. G-protein]. This conformational change is affected by the aforementioned deviances from Class A GPCRs. Rather than a large conformational change, a subtle one is induced. This will allow MRGPRX2 to interact with a G-protein. <scene name='90/904327/Gproteins/2'>G-proteins</scene> are composed of 3 subunits | + | Once the ligand is bound, MRGPRX2 undergoes a conformational change that is transmitted through to the [https://en.wikipedia.org/wiki/G_protein#:~:text=G%20proteins%2C%20also%20known%20as,a%20cell%20to%20its%20interior. G-protein]. This conformational change is affected by the aforementioned deviances from Class A GPCRs. Rather than a large conformational change, a subtle one is induced. This will allow MRGPRX2 to interact with a G-protein. <scene name='90/904327/Gproteins/2'>G-proteins</scene> are composed of 3 subunits: α, β, and γ. When activated, the receptor acts as a [https://en.wikipedia.org/wiki/Guanine_nucleotide_exchange_factor Guanine nucleotide factor (GEF)]which will allow the Gα subunit to have its GDP be replaced by a GTP. This will cause the Gα subunit to dissociate from the dimer Gβγ <ref name="Ramesh">Ramesh, Soliman, et al. (2015) "G-Protein Coupled Receptors (GPCRs): A Comprehensive Computational Perspective." Combinational Chemistry and High Throughout Screening, 18(4), 346-364, https://pubmed.ncbi.nlm.nih.gov/25747435/</ref>. They are characterized by conserved motifs including DRY, PIF, Sodium Binding, and the CWxP <ref name="Zhou">PMID: 31855179</ref>. The Gα subunit is then able to act as a secondary messenger to begin the signal transduction in the cell. MRGPRX2 interacts with two different types of G-proteins; Gi and Gq. These G-proteins are activated by similar interactions with the receptor however, they are structurally different and changes in the induced conformation are observed (Figure 5). |
=== 3.After G-Protein Activation === | === 3.After G-Protein Activation === | ||
Revision as of 13:40, 21 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Itch Mas-Related G-Protein Coupled Receptor
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 1.9 Cao, Can, et al. "Structure, function and pharmacology of human itch GPCRs." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04126-6
- ↑ Thal, David M., et al. "Structural insights into G-protein-coupled receptor allostery." Nature, Nature Publishing Group, 04 July 2018, https://www.nature.com/articles/s41586-018-0259-z
- ↑ 3.0 3.1 Zhang D, Zhao Q, Wu B. Structural Studies of G Protein-Coupled Receptors. Mol Cells. 2015 Oct;38(10):836-42. doi: 10.14348/molcells.2015.0263. Epub 2015, Oct 15. PMID:26467290 doi:http://dx.doi.org/10.14348/molcells.2015.0263
- ↑ 4.0 4.1 Ramesh, Soliman, et al. (2015) "G-Protein Coupled Receptors (GPCRs): A Comprehensive Computational Perspective." Combinational Chemistry and High Throughout Screening, 18(4), 346-364, https://pubmed.ncbi.nlm.nih.gov/25747435/
- ↑ 5.0 5.1 5.2 5.3 Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI, Liu ZJ, Stevens RC, Lambert NA, Babu MM, Wang MW, Zhao S. Common activation mechanism of class A GPCRs. Elife. 2019 Dec 19;8. pii: 50279. doi: 10.7554/eLife.50279. PMID:31855179 doi:http://dx.doi.org/10.7554/eLife.50279
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y
- ↑ 7.0 7.1 Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x
- ↑ Sandoval, A., et al. "The Molecular Switching Mechanism at the Conserved D(E)RY Motif in Class-A GPCRs." Biophysical journal, 111(1), 79-89. https://doi.org/10.1016/j.bpj.2016.06.004
- ↑ Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014 May;39(5):233-44. doi: 10.1016/j.tibs.2014.03.002. Epub , 2014 Apr 21. PMID:24767681 doi:http://dx.doi.org/10.1016/j.tibs.2014.03.002
- ↑ Babina, M., et al. "MRGPRX2 Is the Codeine Receptor of Human Skin Mast Cells: Desensitization through β-Arrestin and Lack of Correlation with the FcεRI Pathway." Journal of Investigative Dermatology, 141(6), 1286-1296. https://doi.org/10.1016/j.jid.2020.09.017
- ↑ McNeil, B. D., et al. "MRGPRX2 and Adverse Drug Reactions." Frontier Immunology, 06 August 2021, https://www.frontiersin.org/articles/10.3389/fimmu.2021.676354/full
- ↑ Ogasawara, H., et al. "Novel MRGPRX2 antagonists inhibit IgE-independent activation of human umbilical cord blood-derived mast cells." Journal of Leukocyte Biology, 12 July 2019, https://jlb.onlinelibrary.wiley.com/doi/10.1002/JLB.2AB1018-405R