This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
LCE1a2
From Proteopedia
(Difference between revisions)
(New page: ==Structure== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for your page '''LCE1a2'''. Click above on '''edit...) |
|||
| Line 5: | Line 5: | ||
== Background == | == Background == | ||
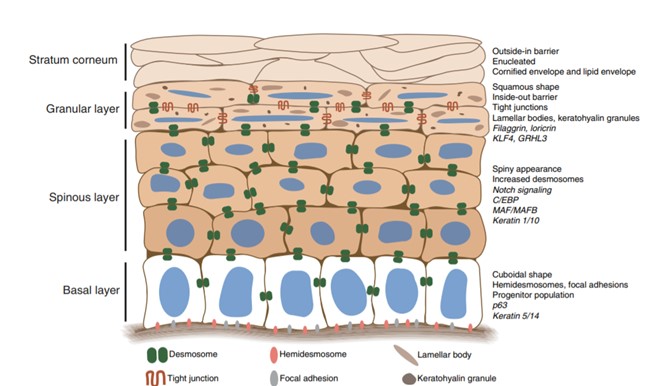
| - | The largest organ of the human body is the skin. The skin is capable of many things which protect us from the outside environment, and it is composed of many layers. The epidermis is the top layer of skin and composed within it are 4 layers: Spinous layer, Granular layer, Stratum corneum, and the Basal layer (''see figure 1''[[Image: | + | The largest organ of the human body is the skin. The skin is capable of many things which protect us from the outside environment, and it is composed of many layers. The epidermis is the top layer of skin and composed within it are 4 layers: Spinous layer, Granular layer, Stratum corneum, and the Basal layer (''see figure 1'' [[Image:Skin.jpg]]). All of these aspects help regulate the epidermal layer. Regulation of the epidermal layer Is essential in reducing inflammatory skin diseases and it’s the first line of defense against unwanted outsiders. The components of the epidermal layer help to ensure that inflammatory aspects and other skin conditions don’t arise such as Psoriasis. Major reasons that conditions arise is due to an irregulating of the skin. |
When it comes to skin “within the epidermal differentiation complex on human chromosome one (mouse chromosome three), [there] contains multiple conserved gene encoding stratum-corneum proteins”; these genes are in a cluster termed LCE’s or Late Cornifying Envelopes (Jackson 2005). These envelopes are composed of 18 genes within chromosome 1 of the human genome and chromosome 3 of mice genomes. Although their function isn’t completely understood, it has been hypothesized that their main function is as an antimicrobial asset to the body. Given that their true purpose is not known yet, the antimicrobial hypothesis is constantly being tested and redeveloped. Having an understanding of the function that LCE’s provide will allow for better dermatological care in the future as well as to better help identify why certain conditions rise or fall. | When it comes to skin “within the epidermal differentiation complex on human chromosome one (mouse chromosome three), [there] contains multiple conserved gene encoding stratum-corneum proteins”; these genes are in a cluster termed LCE’s or Late Cornifying Envelopes (Jackson 2005). These envelopes are composed of 18 genes within chromosome 1 of the human genome and chromosome 3 of mice genomes. Although their function isn’t completely understood, it has been hypothesized that their main function is as an antimicrobial asset to the body. Given that their true purpose is not known yet, the antimicrobial hypothesis is constantly being tested and redeveloped. Having an understanding of the function that LCE’s provide will allow for better dermatological care in the future as well as to better help identify why certain conditions rise or fall. | ||
| Line 15: | Line 15: | ||
In this research specifically, LCE1a2 (found in mice) was looked into to try and create a structural model of it as well as produce a background. Although the specific function of the stratum-corneum proteins that this gene codes for was not determined, the information needed for potential sequencing and future studies was found. The general biological process of this protein is keratinization which was to be expected and further research confirmed it to be a signaling protein for the epidermis. Upon further research the structure of the gene was able to be estimated with 98 percent confidence with respect to the homology of the sequence to the structure formed utilizing Phyr2 programming. To achieve these result the DNA sequence of the LCE1a2 cluster was located via the uniprot page. Once the sequence was found a blast was conducted in order to identify the different proteins that the gene codes for. The protein labeled Late cornified envelope 1A2 Mus musculus (late_cornified_envelope_1A2_[Mus_musculus]) was chosen as it was of the same species and location of the intended search, it was also the highest match probability made by the system. Other proteins produces were late_cornified_envelope_protein_1C-like_[Onychomys_torridus], small_proline_rich-like_2,_partial_[Mus_musculus], small_proline_rich-like_3,_partial_[Mus_musculus], and late_cornified_envelope_1A1_[Mus_musculus]. The reason for the prevalence of small protein rich proteins is due to the reason stated prior which is the fact that LCE’s and SPPR’s are located right next to each other with the genome. As a result, their makeup will more than likely be very similar; due to the high predictive aspects involved with LCE’s currently, it is no surprise that SPRR’s showed up in the protein list as potential matches for the proteins that LCE gene 1a2 codes for. With all of that aside once the protein of intent was identified which was late_cornified_envelope_1A2_[Mus_musculus] it was run and analyzed. Results indicated can be found here. In addition to the protein, a look into the gene itself was done. The DNA sequence of the LCE1a2 gene was obtained and input into an in-silico Gibson assembly cloning method with Laser Gene. Results indicate that the gene has high complexity naturally making the sequencing process slightly more complex as well. The site used to determine complexity can be found here: after getting to the site be sure to click products and services then click costume DNA. This will allow you to view all of the information need to sequence your subject. | In this research specifically, LCE1a2 (found in mice) was looked into to try and create a structural model of it as well as produce a background. Although the specific function of the stratum-corneum proteins that this gene codes for was not determined, the information needed for potential sequencing and future studies was found. The general biological process of this protein is keratinization which was to be expected and further research confirmed it to be a signaling protein for the epidermis. Upon further research the structure of the gene was able to be estimated with 98 percent confidence with respect to the homology of the sequence to the structure formed utilizing Phyr2 programming. To achieve these result the DNA sequence of the LCE1a2 cluster was located via the uniprot page. Once the sequence was found a blast was conducted in order to identify the different proteins that the gene codes for. The protein labeled Late cornified envelope 1A2 Mus musculus (late_cornified_envelope_1A2_[Mus_musculus]) was chosen as it was of the same species and location of the intended search, it was also the highest match probability made by the system. Other proteins produces were late_cornified_envelope_protein_1C-like_[Onychomys_torridus], small_proline_rich-like_2,_partial_[Mus_musculus], small_proline_rich-like_3,_partial_[Mus_musculus], and late_cornified_envelope_1A1_[Mus_musculus]. The reason for the prevalence of small protein rich proteins is due to the reason stated prior which is the fact that LCE’s and SPPR’s are located right next to each other with the genome. As a result, their makeup will more than likely be very similar; due to the high predictive aspects involved with LCE’s currently, it is no surprise that SPRR’s showed up in the protein list as potential matches for the proteins that LCE gene 1a2 codes for. With all of that aside once the protein of intent was identified which was late_cornified_envelope_1A2_[Mus_musculus] it was run and analyzed. Results indicated can be found here. In addition to the protein, a look into the gene itself was done. The DNA sequence of the LCE1a2 gene was obtained and input into an in-silico Gibson assembly cloning method with Laser Gene. Results indicate that the gene has high complexity naturally making the sequencing process slightly more complex as well. The site used to determine complexity can be found here: after getting to the site be sure to click products and services then click costume DNA. This will allow you to view all of the information need to sequence your subject. | ||
Although the direct capabilities of the proteins coded for via the LCE clusters are still not known, the information provided above gives not only background of the topic of LCE’s but also provides look into the structure and properties of a protein from LCE1a2 cluster. In addition to the characteristics of a protein found within the gene cluster, the steps used to find and analyze the protein was given. Alongside the protein a clonal sequence was produced to provide additional sequence characteristics such as melting point and complexity. Those results can be found ''here''. | Although the direct capabilities of the proteins coded for via the LCE clusters are still not known, the information provided above gives not only background of the topic of LCE’s but also provides look into the structure and properties of a protein from LCE1a2 cluster. In addition to the characteristics of a protein found within the gene cluster, the steps used to find and analyze the protein was given. Alongside the protein a clonal sequence was produced to provide additional sequence characteristics such as melting point and complexity. Those results can be found ''here''. | ||
| - | |||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | == Conclusion == | ||
| + | Overall, the topic of late cornifying envelopes is still not fully known much like many other scientific topics; with that said this report provides key background information on the importance, location, proposed capabilities, as well as an idea of specific gene characteristics. With this information in mind further studies can be conducted to try and clone these genes and figure out their individual function and overall capabilities. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 20:17, 23 April 2022
Structure
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644