We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
BASIL2022GV3HDT
From Proteopedia
(Difference between revisions)
| Line 48: | Line 48: | ||
===='''Coupled kinase assay'''==== | ===='''Coupled kinase assay'''==== | ||
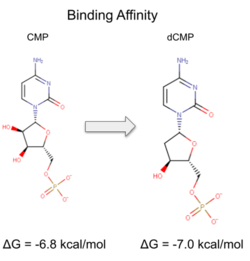
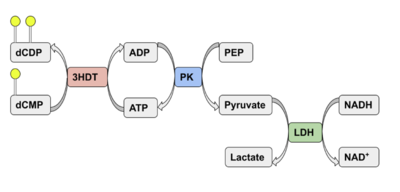
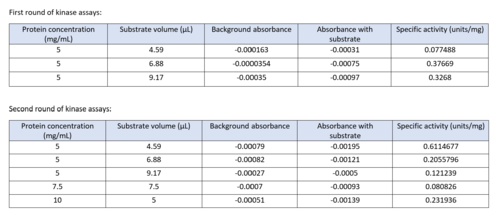
| - | Two rounds of coupled kinase assays were run using 3HDT with ATP and dCMP as substrates. The concentration of dCMP was 109mM. The first round of assay (3 total assays) used 5μL of 3HDT and various amounts of 109mM of dCMP. 6.88μL of dCMP resulted in the highest specific activity (0.37669 U/mg) and increasing the substrate amount ~2μL had a similar but slightly less specific activity of 0.3268 U/mg. However, when we repeated the first kinase assays we did (using 5μL of 3HDT), the results | + | Two rounds of coupled kinase assays were run using 3HDT with ATP and dCMP as substrates. The concentration of dCMP was 109mM. The first round of assay (3 total assays) used 5μL of 3HDT and various amounts of 109mM of dCMP. 6.88μL of dCMP resulted in the highest specific activity (0.37669 U/mg) and increasing the substrate amount ~2μL had a similar but slightly less specific activity of 0.3268 U/mg. However, when we repeated the first kinase assays we did (using 5μL of 3HDT), there are discrepancies in the results indicating a potential experimental error such as not pipetting up and down to mix, bubbles, taking too long between mixing the substrate in and reading the plate. In addition, the protein in the second round was older (original protein but about a week old from when it was made) which could have affected the specific activity with the substrate because the protein was starting to expire/decrease function. |
Revision as of 04:16, 24 April 2022
Characterizing Putative Kinase 3HDT
| |||||||||||
References
1. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
2. National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
3. Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
4. The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky