This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Titin
From Proteopedia
(Difference between revisions)
| Line 40: | Line 40: | ||
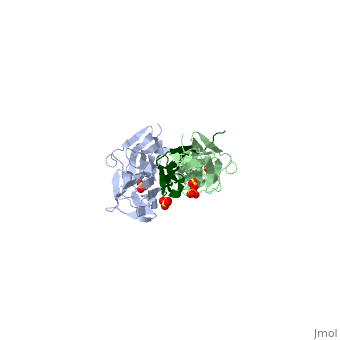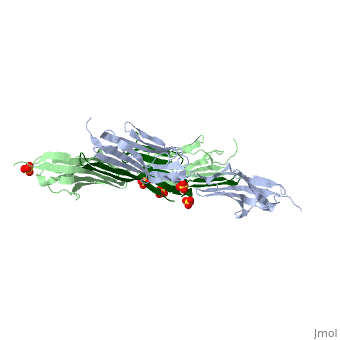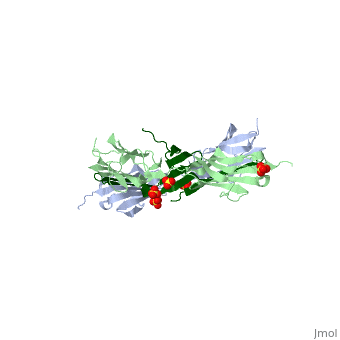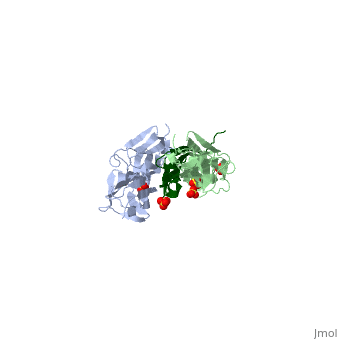
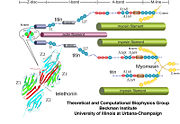
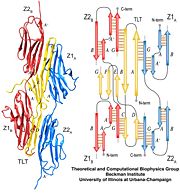
As seen in the image (Z1Z2 / Telethonin complex), the major force enduring component of this complex is an elaborate intermolecular hydrogen bonding network formed across <scene name='2a38/Test/2'>β-strand</scene> among telethonin and Z1Z2 domains, and not intramolecularly among termini β-strands of individual Z1 or Z2 domains. This shift to a stronger force enduring interface reduces the possibility of unraveling the individual Ig-domains, thus stabilizing the complex. This demonstrates how <scene name='2a38/Test/2'>β-strand</scene> cross-linking via [http://en.wikipedia.org/wiki/Hydrogen_bonds hydrogen bonds] serves as an important mechanism. It functions as a molecular adhesive, increasing the ability of protein complexes to resist against mechanical stress. | As seen in the image (Z1Z2 / Telethonin complex), the major force enduring component of this complex is an elaborate intermolecular hydrogen bonding network formed across <scene name='2a38/Test/2'>β-strand</scene> among telethonin and Z1Z2 domains, and not intramolecularly among termini β-strands of individual Z1 or Z2 domains. This shift to a stronger force enduring interface reduces the possibility of unraveling the individual Ig-domains, thus stabilizing the complex. This demonstrates how <scene name='2a38/Test/2'>β-strand</scene> cross-linking via [http://en.wikipedia.org/wiki/Hydrogen_bonds hydrogen bonds] serves as an important mechanism. It functions as a molecular adhesive, increasing the ability of protein complexes to resist against mechanical stress. | ||
| + | == 3D Structures of Titin == | ||
| + | [[Titin 3D structures]] | ||
== 3D Structures of Titin == | == 3D Structures of Titin == | ||
| Line 51: | Line 53: | ||
[[4qeg]], [[5jdj]] - hTTN I10 residues 2880-2967<br /> | [[4qeg]], [[5jdj]] - hTTN I10 residues 2880-2967<br /> | ||
[[1tit]], [[1tiu]]- hTTN I27 residues 5253-5341 – NMR<br /> | [[1tit]], [[1tiu]]- hTTN I27 residues 5253-5341 – NMR<br /> | ||
| + | [[7nip]] - hTTN residues 8578-8617 - NMR<br /> | ||
[[5joe]] - hTTN I81 residues 9582-9671<br /> | [[5joe]] - hTTN I81 residues 9582-9671<br /> | ||
| + | [[7ahs]] - hTTN residues 9582-9851<br /> | ||
[[2rq8]] – hTTN residues 12677-12765 (mutant) – NMR<br /> | [[2rq8]] – hTTN residues 12677-12765 (mutant) – NMR<br /> | ||
[[1waa]] - hTTN residues 12801-12889<br /> | [[1waa]] - hTTN residues 12801-12889<br /> | ||
| Line 63: | Line 67: | ||
[[1nct]], [[1ncu]] – hTTN M5 residues 26059-26155 – NMR<br /> | [[1nct]], [[1ncu]] – hTTN M5 residues 26059-26155 – NMR<br /> | ||
[[1tnm]], [[1tnn]] – hTTN M5 residues 26059-26155<br /> | [[1tnm]], [[1tnn]] – hTTN M5 residues 26059-26155<br /> | ||
| + | [[6ygn]] – hTTN residues 30596-31144<br /> | ||
[[3lcy]] – hTTN A164-A165 residues 31456-31649<br /> | [[3lcy]] – hTTN A164-A165 residues 31456-31649<br /> | ||
[[2nzi]] – hTTN residues 31854-32155<br /> | [[2nzi]] – hTTN residues 31854-32155<br /> | ||
| Line 77: | Line 82: | ||
[[3b43]] - rTTN I65-I70<br /> | [[3b43]] - rTTN I65-I70<br /> | ||
[[1h8b]] – rTTN residues 648-698 + hα Actinin 2 EF hands 3&4<br /> | [[1h8b]] – rTTN residues 648-698 + hα Actinin 2 EF hands 3&4<br /> | ||
| + | [[6yj0]] - mTTN I83 (mutant) residues 9719-9809 - mouse<br /> | ||
[[6i0y]] – TTN I27 residues 14318-14406 in 50S ribosome – ''Escherichia coli'' – Cryo EM<br /> | [[6i0y]] – TTN I27 residues 14318-14406 in 50S ribosome – ''Escherichia coli'' – Cryo EM<br /> | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 08:56, 25 April 2022
| |||||||||||
References
- http://www.ncbi.nlm.nih.gov:80/pmc/articles/PMC1948054/?tool=pmcentrez
- http://www.ks.uiuc.edu/Research/z1z2/
- http://www.ks.uiuc.edu/Research/telethonin/
- http://de.wikipedia.org/wiki/Titin
Created with the participation of Anton Schmidt, Wolfgang Hermann.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Jaime Prilusky
Categories: Topic Page | Homo sapiens | Demirel, M. | Marino, M. | Mayans, O. | Muhle-Goll, C. | Svergun, D. | Titin | Z1z2