BASIL2022GV3HDT
From Proteopedia
| Line 20: | Line 20: | ||
===='''Pfam'''==== | ===='''Pfam'''==== | ||
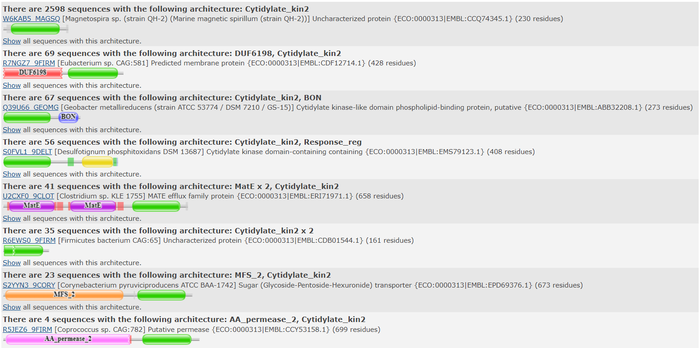
| - | We used the FASTA sequence from 3HDT to do a comparative search in Pfam<ref> | + | We used the FASTA sequence from 3HDT to do a comparative search in Pfam<ref>Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman |
| + | Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913</ref> to find similar protein families our protein may belong. Pfam predicted that our protein is part of the cytidylate kinase which was also shown in the BLASTp results. | ||
[[Image:Pfam search resultsJRAF2022.png |800px| center | thumb | Pfam query aligned with a cytidylate kinase-like family.]] | [[Image:Pfam search resultsJRAF2022.png |800px| center | thumb | Pfam query aligned with a cytidylate kinase-like family.]] | ||
| Line 28: | Line 29: | ||
===='''DALI'''==== | ===='''DALI'''==== | ||
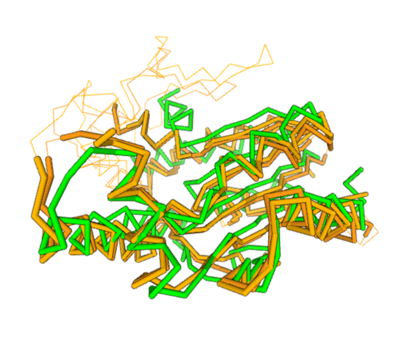
| - | Two of the top results from our DALI query (both cytidylate kinases) show a high structural resemblance with 3hdt when overlaid in DALI’s viewer. It is also worth noting that the majority of our results from our DALI query consisted of cytidylate kinases. This information helped our decision to choose a function like cytidylate kinase because of similar alignment between the proteins (structure of the protein = function) indicating similarity in function and our protein relates to them. | + | Two of the top results from our DALI<ref>Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.</ref> query (both cytidylate kinases) show a high structural resemblance with 3hdt when overlaid in DALI’s viewer. It is also worth noting that the majority of our results from our DALI query consisted of cytidylate kinases. This information helped our decision to choose a function like cytidylate kinase because of similar alignment between the proteins (structure of the protein = function) indicating similarity in function and our protein relates to them. |
| Line 35: | Line 36: | ||
== Docking== | == Docking== | ||
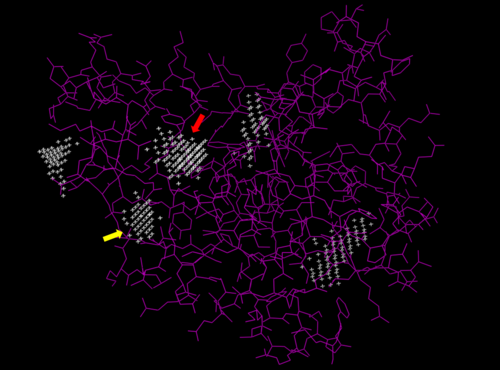
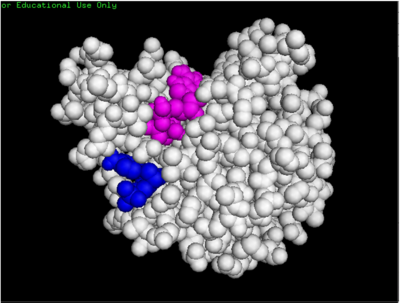
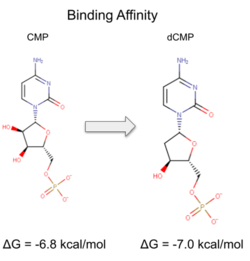
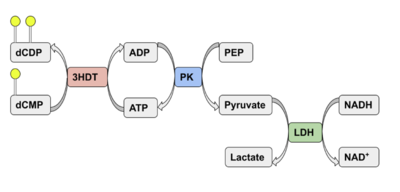
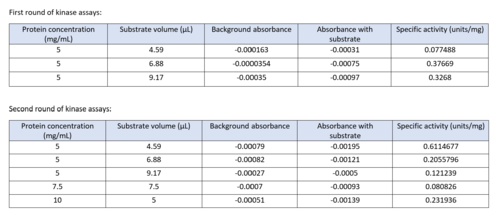
| - | We used POCASA to determine potential binding pockets within our protein and PyRx to actually bind dCMP to 3HDT. Then PyMOL was used to visualize the binding pockets and dCMP in the protein. This <scene name='90/904996/Binding_pockets/1'>binding pocket</scene> (in purple) is a potential pocket the substrate dCMP may bind to in the protein, 3HDT. However, this area was where dCMP binded with the highest affinity in PyRx. The amino acids interacting with the substrate within that area are.... | + | We used POCASA to determine potential binding pockets within our protein and PyRx<ref>Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.</ref> to actually bind dCMP to 3HDT. Then PyMOL was used to visualize the binding pockets and dCMP in the protein. This <scene name='90/904996/Binding_pockets/1'>binding pocket</scene> (in purple) is a potential pocket the substrate dCMP may bind to in the protein, 3HDT. However, this area was where dCMP binded with the highest affinity in PyRx. The amino acids interacting with the substrate within that area are.... |
However, these results may not be as accurate because during the docking process on PyRx, we were unsuccessful | However, these results may not be as accurate because during the docking process on PyRx, we were unsuccessful | ||
Revision as of 04:20, 26 April 2022
Characterizing Putative Kinase 3HDT
| |||||||||||
References
- ↑ National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
- ↑ Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
- ↑ Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
- ↑ Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.
1. National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; [1988] – [cited 2022 April 23].
2. Pfam: The protein families database in 2021: J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu Schrödinger, LLC.
5. Small-Molecule Library Screening by Docking with PyRx. Dallakyan S, Olson AJ. Methods Mol Biol. 2015;1263:243-50.
Proteopedia Page Contributors and Editors (what is this?)
Jesse D. Rothfus, Autumn Forrester, Bonnie Hall, Jaime Prilusky