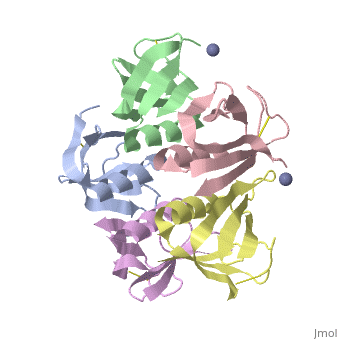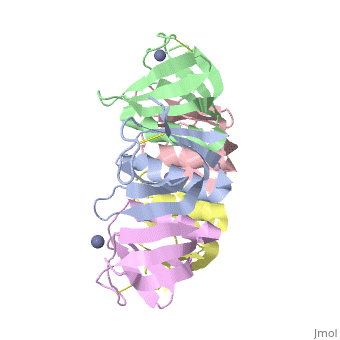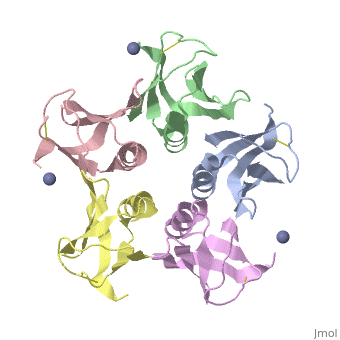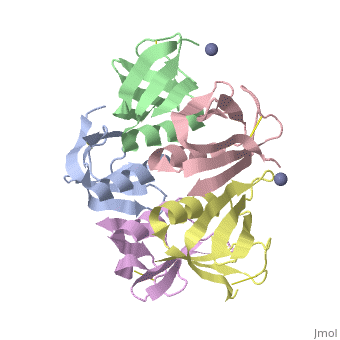2xsc
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='2xsc' size='340' side='right'caption='[[2xsc]], [[Resolution|resolution]] 2.05Å' scene=''> | <StructureSection load='2xsc' size='340' side='right'caption='[[2xsc]], [[Resolution|resolution]] 2.05Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2xsc]] is a 5 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[2xsc]] is a 5 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1bov 1bov]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2XSC OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2XSC FirstGlance]. <br> |
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | ||
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1ojf|1ojf]]</td></tr> | + | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[1ojf|1ojf]]</div></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2xsc FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2xsc OCA], [https://pdbe.org/2xsc PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2xsc RCSB], [https://www.ebi.ac.uk/pdbsum/2xsc PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2xsc ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [[ | + | [[https://www.uniprot.org/uniprot/STXB_BPH30 STXB_BPH30]] The B subunit is responsible for the binding of the holotoxin to specific receptors on the target cell surface, such as globotriaosylceramide (Gb3) in human intestinal microvilli. |
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
== Publication Abstract from PubMed == | == Publication Abstract from PubMed == | ||
| Line 22: | Line 22: | ||
==See Also== | ==See Also== | ||
*[[Shiga toxin|Shiga toxin]] | *[[Shiga toxin|Shiga toxin]] | ||
| + | *[[Shiga toxin 3D structures|Shiga toxin 3D structures]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 12:16, 27 April 2022
Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli
| |||||||||||