Serine hydroxymethyltransferase
From Proteopedia
(Difference between revisions)
| Line 44: | Line 44: | ||
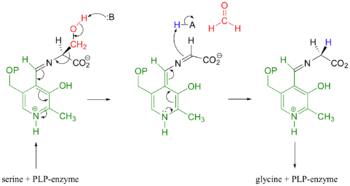
[[Image:S_to_G_Mechanism.png|right|thumb|350px| Mechanistic Conversion of Serine to Glycine catalyzed by SHMT]] | [[Image:S_to_G_Mechanism.png|right|thumb|350px| Mechanistic Conversion of Serine to Glycine catalyzed by SHMT]] | ||
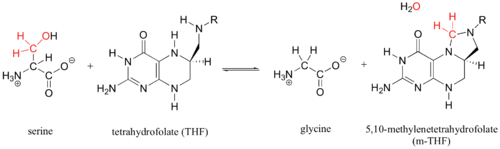
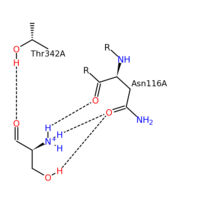
| - | The most widely accepted mechanism of action is through the process of modified retro-aldol cleavage. In this case, the mechanism starts with the serine/PLP-enzyme complex. The base represented by 'B' attacks the hydrogen of the hydroxyl group on serine. Resonance allows the process of tautomerization to occur. This end results of tautomerization results in the formation of a formaldehyde, and acidic 'H-A' compound, and an imine attached to the serine-PLP complex. The double bond of the imine then reacts with the acidic 'H-A' compound. Resonance then allows the new complex to form glycine and the PLP-enzyme.<ref>PMID:123456</ref> | + | The most widely accepted mechanism of action is through the process of modified retro-aldol cleavage. In this case, the mechanism starts with the serine/PLP-enzyme complex. The base represented by 'B' attacks the hydrogen of the hydroxyl group on serine. Resonance allows the process of tautomerization to occur. This end results of tautomerization results in the formation of a formaldehyde, and acidic 'H-A' compound, and an imine attached to the serine-PLP complex. The double bond of the imine then reacts with the acidic 'H-A' compound. Resonance then allows the new complex to form glycine and the PLP-enzyme.<ref>PMID:123456</ref><ref>DOI:10.1021/acscatal.8b02321</ref>. An alternative scheme shows direct transfer of the hydroxymethyl group from serine to tetrahydrofolate via a covalent bisubstrate intermediate (see also [[Thymidylate synthase]]). In the crystal structure of a ternary complex of glycine (covalently linked to PLP) and N5-formyltetrahydrofolate, the two intermediates are in <scene name='56/566522/Methyl_transfer/1'>close proximity</scene>.<ref>DOI:10.1074/jbc.M111976200</ref>. |
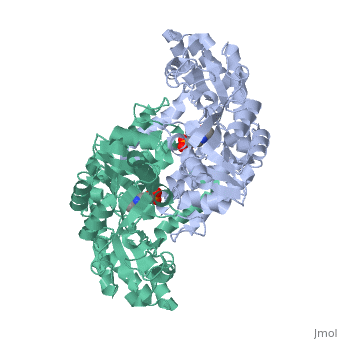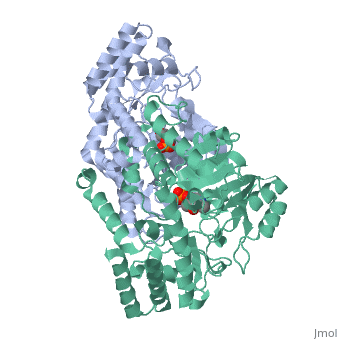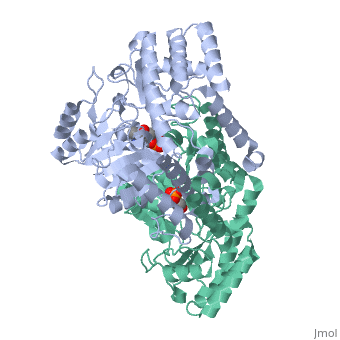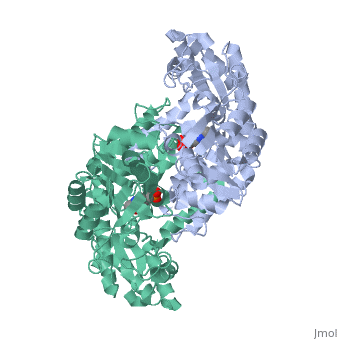
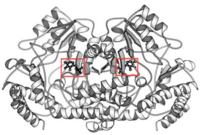
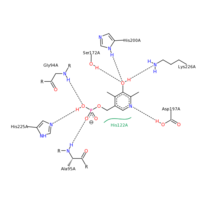
In the case of serine hydroxymethyltransferase, the pyridoxal-5'-phosphate is the cofactor. The SHMT enzyme has no enzymatic activity without the presence of this cofactor. On the large C-terminal domain, PLP is covalently bound to the N-terminus of an alpha helix following a beta strand. The K226 residue specifically is important for the binding. The amino group on this lysine residue helps bind PLP. The phosphate group of PLP is bound to the N-terminus of the next alpha helix. The PLP will pack against the B strands present. The binding of PLP to SHMT supports conformation of the active site which is located in a space between the two monomeric domains at the border between the two monomeric units of the SHMT dimer. For now, serine and tetrahydrofolate (THF) are the only known biological substrates of SHMT. The end products are of course glycine and 5,10-methylene tetrahydrofolate. If the reaction is flipped, the substrates and products are also flipped. An important point of the SHMT mechanism is that fact that the reaction can start with either serine or glycine as the substrate. As far as inhibitors, antifolates are known to reduce and/or eliminate the enzymatic activity of SHMT. This is not surprising because of the fact that folate compounds are substrates of SHMT.<ref>PMID:123456</ref><ref>PMID:16125438</ref> | In the case of serine hydroxymethyltransferase, the pyridoxal-5'-phosphate is the cofactor. The SHMT enzyme has no enzymatic activity without the presence of this cofactor. On the large C-terminal domain, PLP is covalently bound to the N-terminus of an alpha helix following a beta strand. The K226 residue specifically is important for the binding. The amino group on this lysine residue helps bind PLP. The phosphate group of PLP is bound to the N-terminus of the next alpha helix. The PLP will pack against the B strands present. The binding of PLP to SHMT supports conformation of the active site which is located in a space between the two monomeric domains at the border between the two monomeric units of the SHMT dimer. For now, serine and tetrahydrofolate (THF) are the only known biological substrates of SHMT. The end products are of course glycine and 5,10-methylene tetrahydrofolate. If the reaction is flipped, the substrates and products are also flipped. An important point of the SHMT mechanism is that fact that the reaction can start with either serine or glycine as the substrate. As far as inhibitors, antifolates are known to reduce and/or eliminate the enzymatic activity of SHMT. This is not surprising because of the fact that folate compounds are substrates of SHMT.<ref>PMID:123456</ref><ref>PMID:16125438</ref> | ||
Revision as of 21:22, 30 April 2022
| |||||||||||
References
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Elsea SH, Juyal RC, Jiralerspong S, Finucane BM, Pandolfo M, Greenberg F, Baldini A, Stover P, Patel PI. Haploinsufficiency of cytosolic serine hydroxymethyltransferase in the Smith-Magenis syndrome. Am J Hum Genet. 1995 Dec;57(6):1342-50. PMID:8533763
- ↑ Renwick SB, Snell K, Baumann U. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998 Sep 15;6(9):1105-16. PMID:9753690
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Grados OB. [The laboratory in programs for enteric infection control]. Bol Oficina Sanit Panam. 1975 Apr;78(4):318-22. PMID:123456
- ↑ doi: https://dx.doi.org/10.1021/acscatal.8b02321
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Grados OB. [The laboratory in programs for enteric infection control]. Bol Oficina Sanit Panam. 1975 Apr;78(4):318-22. PMID:123456
- ↑ Schirch V, Szebenyi DM. Serine hydroxymethyltransferase revisited. Curr Opin Chem Biol. 2005 Oct;9(5):482-7. PMID:16125438 doi:http://dx.doi.org/10.1016/j.cbpa.2005.08.017
- ↑ Renwick SB, Snell K, Baumann U. The crystal structure of human cytosolic serine hydroxymethyltransferase: a target for cancer chemotherapy. Structure. 1998 Sep 15;6(9):1105-16. PMID:9753690
- ↑ Trivedi V, Gupta A, Jala VR, Saravanan P, Rao GS, Rao NA, Savithri HS, Subramanya HS. Crystal structure of binary and ternary complexes of serine hydroxymethyltransferase from Bacillus stearothermophilus: insights into the catalytic mechanism. J Biol Chem. 2002 May 10;277(19):17161-9. Epub 2002 Feb 27. PMID:11877399 doi:10.1074/jbc.M111976200
- ↑ Elsea SH, Juyal RC, Jiralerspong S, Finucane BM, Pandolfo M, Greenberg F, Baldini A, Stover P, Patel PI. Haploinsufficiency of cytosolic serine hydroxymethyltransferase in the Smith-Magenis syndrome. Am J Hum Genet. 1995 Dec;57(6):1342-50. PMID:8533763
Proteopedia Page Contributors and Editors (what is this?)
Coleman Calva, Michal Harel, Karsten Theis, Alexander Berchansky, Jaime Prilusky