User:Kiera Malone/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
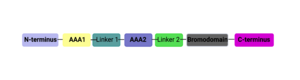
==The ATPase Family, AAA Domain-Containing Protein 2B '''(ATAD2B)'''== | ==The ATPase Family, AAA Domain-Containing Protein 2B '''(ATAD2B)'''== | ||
| - | <StructureSection load='3lxj' size='350' side='right' caption= | + | <StructureSection load='3lxj' size='350' side='right' caption='ATAD2B bromodomain' scene=''> |
== Introduction == | == Introduction == | ||
| Line 92: | Line 92: | ||
===Compound 38=== | ===Compound 38=== | ||
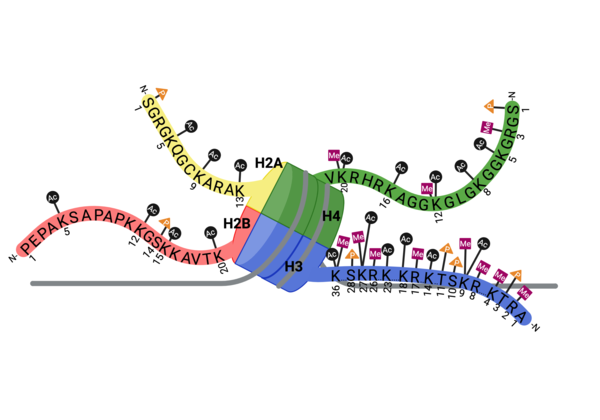
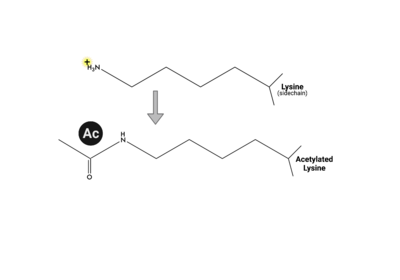
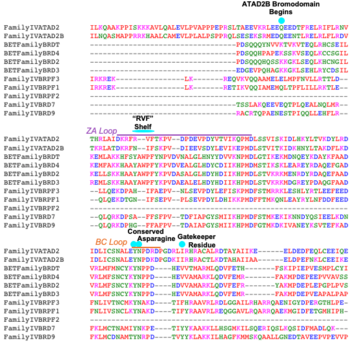
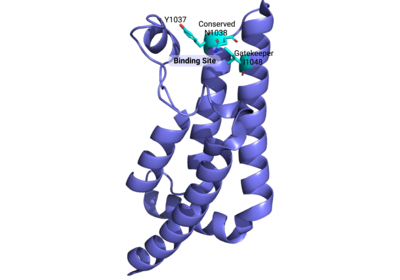
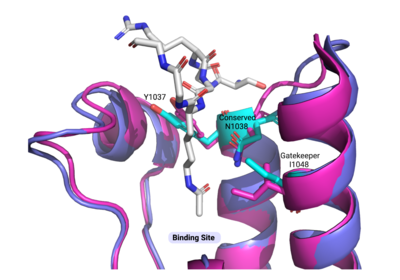
Compound 38 (C-38) was 1.8-fold times more selective for the ATAD2 <font color='gray'>bromodomain</font> than ATAD2B, with 90 nM and 166.3 nM binding affinities, respectively. They solved the <scene name='90/909366/C38/1'>structure</scene> of C-38 bound to the ATAD2B <font color='gray'>bromodomain</font> to determine the specific molecular interactions that are occurring (PDB ID: 6VEO). C-38 was coordinated in the binding pocket much like the acetylated lysine residues of the histone proteins: through hydrogen bonds and hydrophobic interactions. The inhibitor bound to the conserved asparagine (N1038), and was coordinated through hydrophobic interactions with the gatekeeper residue I1048. However, since this compound was manufactured to be specific for the ATAD2 <font color='gray'>bromodomain</font>, some important interactions between protein and inhibitor are not present in the ATAD2B <font color='gray'>bromodomain</font> structure, including an important hydrogen bond interaction. These mechanistic molecular differences are thought to play a role in the lower affinity for C-38 with ATAD2B. Overall, however, the coordination of C-38 between ATAD2 and ATAD2B is extremely similar, and Lloyd, et al.<ref>PMID:33084328</ref> postulates that it may be difficult to develop a truly selective inhibitor for each protein. This endeavor is still being explored. | Compound 38 (C-38) was 1.8-fold times more selective for the ATAD2 <font color='gray'>bromodomain</font> than ATAD2B, with 90 nM and 166.3 nM binding affinities, respectively. They solved the <scene name='90/909366/C38/1'>structure</scene> of C-38 bound to the ATAD2B <font color='gray'>bromodomain</font> to determine the specific molecular interactions that are occurring (PDB ID: 6VEO). C-38 was coordinated in the binding pocket much like the acetylated lysine residues of the histone proteins: through hydrogen bonds and hydrophobic interactions. The inhibitor bound to the conserved asparagine (N1038), and was coordinated through hydrophobic interactions with the gatekeeper residue I1048. However, since this compound was manufactured to be specific for the ATAD2 <font color='gray'>bromodomain</font>, some important interactions between protein and inhibitor are not present in the ATAD2B <font color='gray'>bromodomain</font> structure, including an important hydrogen bond interaction. These mechanistic molecular differences are thought to play a role in the lower affinity for C-38 with ATAD2B. Overall, however, the coordination of C-38 between ATAD2 and ATAD2B is extremely similar, and Lloyd, et al.<ref>PMID:33084328</ref> postulates that it may be difficult to develop a truly selective inhibitor for each protein. This endeavor is still being explored. | ||
| + | |||
| + | ==Conclusion== | ||
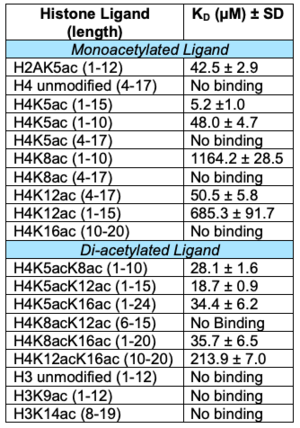
| + | Overall, more information is needed in order to understand the true biological function of ATAD2B. Due to bromodomains being conserved throughout evolution and after being divided into eight sub-families, it is fortunate to have similar proteins to study the structure and function of ATAD2B through. We know that the bromodomain can recognize mono- and di-acetylated histone proteins with micromolar affinity, and that the most important binding residues align well with the ATAD2 bromodomain, despite there not being a structure for ATAD2B bound to a histone protein. | ||
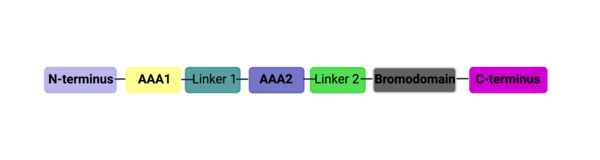
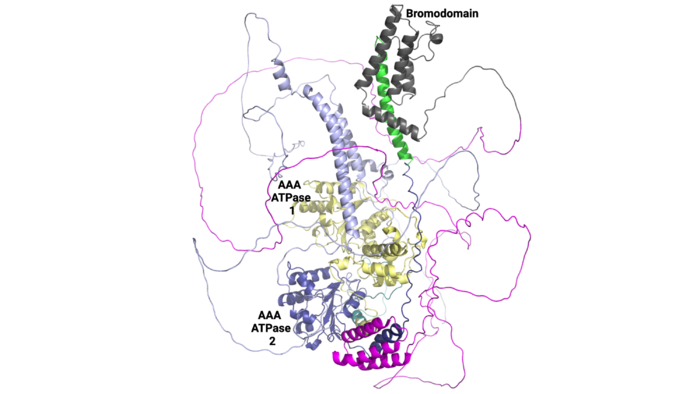
| + | AlphaFold helps us to visualize the entire length of the ATAD2B protein. While interesting and insightful, there are many loop regions, and we do not know what the function of the other domains are yet. It would be interesting to investigate ATAD2B further to determine more about its function so that we can learn from its structure. | ||
| + | |||
| + | There are many players in the story and function of ATAD2B, but little knowledge. In the future, it will be interesting to see what new insights develop into its structure and function. | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 03:23, 2 May 2022
The ATPase Family, AAA Domain-Containing Protein 2B (ATAD2B)
| |||||||||||
References
- ↑ Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010 Dec;52(9):747-55. doi: 10.1111/j.1440-169X.2010.01211.x. PMID:21158754 doi:http://dx.doi.org/10.1111/j.1440-169X.2010.01211.x
- ↑ Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, Rousseaux S, Khochbin S. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010 Sep 16;29(37):5171-81. doi: 10.1038/onc.2010.259. Epub 2010 Jun, 28. PMID:20581866 doi:http://dx.doi.org/10.1038/onc.2010.259
- ↑ Kalashnikova EV, Revenko AS, Gemo AT, Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD, Chen HW. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010 Nov 15;70(22):9402-12. doi: 10.1158/0008-5472.CAN-10-1199. Epub , 2010 Sep 23. PMID:20864510 doi:http://dx.doi.org/10.1158/0008-5472.CAN-10-1199
- ↑ Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, Helin K. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009 Nov 1;69(21):8491-8. doi: 10.1158/0008-5472.CAN-09-2131. Epub, 2009 Oct 20. PMID:19843847 doi:http://dx.doi.org/10.1158/0008-5472.CAN-09-2131
- ↑ Ciro M, Prosperini E, Quarto M, Grazini U, Walfridsson J, McBlane F, Nucifero P, Pacchiana G, Capra M, Christensen J, Helin K. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009 Nov 1;69(21):8491-8. doi: 10.1158/0008-5472.CAN-09-2131. Epub, 2009 Oct 20. PMID:19843847 doi:http://dx.doi.org/10.1158/0008-5472.CAN-09-2131
- ↑ Leachman NT, Brellier F, Ferralli J, Chiquet-Ehrismann R, Tucker RP. ATAD2B is a phylogenetically conserved nuclear protein expressed during neuronal differentiation and tumorigenesis. Dev Growth Differ. 2010 Dec;52(9):747-55. doi: 10.1111/j.1440-169X.2010.01211.x. PMID:21158754 doi:http://dx.doi.org/10.1111/j.1440-169X.2010.01211.x
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007 Aug 13;26(37):5521-7. doi: 10.1038/sj.onc.1210618. PMID:17694091 doi:http://dx.doi.org/10.1038/sj.onc.1210618
- ↑ Evans CM, Phillips M, Malone KL, Tonelli M, Cornilescu G, Cornilescu C, Holton SJ, Gorjanacz M, Wang L, Carlson S, Gay JC, Nix JC, Demeler B, Markley JL, Glass KC. Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain. Int J Mol Sci. 2021 Aug 24;22(17). pii: ijms22179128. doi: 10.3390/ijms22179128. PMID:34502039 doi:http://dx.doi.org/10.3390/ijms22179128
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178
- ↑ Cho C, Jang J, Kang Y, Watanabe H, Uchihashi T, Kim SJ, Kato K, Lee JY, Song JJ. Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone. Nat Commun. 2019 Dec 17;10(1):5764. doi: 10.1038/s41467-019-13743-9. PMID:31848341 doi:http://dx.doi.org/10.1038/s41467-019-13743-9
- ↑ Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999 Oct 15;286(5439):481-6. doi: 10.1126/science.286.5439.481. PMID:10521337 doi:http://dx.doi.org/10.1126/science.286.5439.481
- ↑ Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019 Jul;571(7766):489-499. doi: 10.1038/s41586-019-1411-0. Epub 2019 Jul, 24. PMID:31341302 doi:http://dx.doi.org/10.1038/s41586-019-1411-0
- ↑ Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41-5. doi: 10.1038/47412. PMID:10638745 doi:http://dx.doi.org/10.1038/47412
- ↑ Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997 Sep 18;389(6648):251-60. PMID:9305837 doi:10.1038/38444
- ↑ Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41-5. doi: 10.1038/47412. PMID:10638745 doi:http://dx.doi.org/10.1038/47412
- ↑ Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008 Jul;40(7):897-903. doi: 10.1038/ng.154. Epub 2008 Jun 15. PMID:18552846 doi:http://dx.doi.org/10.1038/ng.154
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178
- ↑ Koo SJ, Fernandez-Montalvan AE, Badock V, Ott CJ, Holton SJ, von Ahsen O, Toedling J, Vittori S, Bradner JE, Gorjanacz M. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016 Oct 25;7(43):70323-70335. doi: 10.18632/oncotarget.11855. PMID:27612420 doi:http://dx.doi.org/10.18632/oncotarget.11855
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178
- ↑ Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Jul 15. pii: 10.1038/s41586-021-03819-2. doi:, 10.1038/s41586-021-03819-2. PMID:34265844 doi:http://dx.doi.org/10.1038/s41586-021-03819-2
- ↑ Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Zidek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022 Jan 7;50(D1):D439-D444. doi: 10.1093/nar/gkab1061. PMID:34791371 doi:http://dx.doi.org/10.1093/nar/gkab1061
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, Rousseaux S, Khochbin S. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010 Sep 16;29(37):5171-81. doi: 10.1038/onc.2010.259. Epub 2010 Jun, 28. PMID:20581866 doi:http://dx.doi.org/10.1038/onc.2010.259
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18067-72. Epub 2007 Nov 12. PMID:17998543 doi:http://dx.doi.org/10.1073/pnas.0705814104
- ↑ Koo SJ, Fernandez-Montalvan AE, Badock V, Ott CJ, Holton SJ, von Ahsen O, Toedling J, Vittori S, Bradner JE, Gorjanacz M. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016 Oct 25;7(43):70323-70335. doi: 10.18632/oncotarget.11855. PMID:27612420 doi:http://dx.doi.org/10.18632/oncotarget.11855
- ↑ Caron C, Lestrat C, Marsal S, Escoffier E, Curtet S, Virolle V, Barbry P, Debernardi A, Brambilla C, Brambilla E, Rousseaux S, Khochbin S. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene. 2010 Sep 16;29(37):5171-81. doi: 10.1038/onc.2010.259. Epub 2010 Jun, 28. PMID:20581866 doi:http://dx.doi.org/10.1038/onc.2010.259
- ↑ Bamborough P, Chung CW, Demont EH, Furze RC, Bannister AJ, Che KH, Diallo H, Douault C, Grandi P, Kouzarides T, Michon AM, Mitchell DJ, Prinjha RK, Rau C, Robson S, Sheppard RJ, Upton R, Watson RJ. A Chemical Probe for the ATAD2 Bromodomain. Angew Chem Int Ed Engl. 2016 Sep 12;55(38):11382-6. doi: 10.1002/anie.201603928. , Epub 2016 Aug 17. PMID:27530368 doi:http://dx.doi.org/10.1002/anie.201603928
- ↑ Bamborough P, Chung CW, Demont EH, Furze RC, Bannister AJ, Che KH, Diallo H, Douault C, Grandi P, Kouzarides T, Michon AM, Mitchell DJ, Prinjha RK, Rau C, Robson S, Sheppard RJ, Upton R, Watson RJ. A Chemical Probe for the ATAD2 Bromodomain. Angew Chem Int Ed Engl. 2016 Sep 12;55(38):11382-6. doi: 10.1002/anie.201603928. , Epub 2016 Aug 17. PMID:27530368 doi:http://dx.doi.org/10.1002/anie.201603928
- ↑ Demont EH, Chung CW, Furze RC, Grandi P, Michon AM, Wellaway C, Barrett N, Bridges AM, Craggs PD, Diallo H, Dixon DP, Douault C, Emmons AJ, Jones EJ, Karamshi BV, Locke K, Mitchell DJ, Mouzon BH, Prinjha RK, Roberts AD, Sheppard RJ, Watson RJ, Bamborough P. Fragment-Based Discovery of Low-Micromolar ATAD2 Bromodomain Inhibitors. J Med Chem. 2015 Jul 9. PMID:26155854 doi:http://dx.doi.org/10.1021/acs.jmedchem.5b00772
- ↑ Yao D, Zhang J, Wang J, Pan D, He Z. Discovery of novel ATAD2 bromodomain inhibitors that trigger apoptosis and autophagy in breast cells by structure-based virtual screening. J Enzyme Inhib Med Chem. 2020 Dec;35(1):713-725. doi:, 10.1080/14756366.2020.1740924. PMID:32174193 doi:http://dx.doi.org/10.1080/14756366.2020.1740924
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178
- ↑ Lloyd JT, McLaughlin K, Lubula MY, Gay JC, Dest A, Gao C, Phillips M, Tonelli M, Cornilescu G, Marunde MR, Evans CM, Boyson SP, Carlson S, Keogh MC, Markley JL, Frietze S, Glass KC. Structural Insights into the Recognition of Mono- and Diacetylated Histones by the ATAD2B Bromodomain. J Med Chem. 2020 Oct 21. doi: 10.1021/acs.jmedchem.0c01178. PMID:33084328 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c01178