Ceramidase
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
| - | |||
| - | Refs <ref name="Okino">PMID:9603946</ref>, <ref name="Inoue">PMID:19088069</ref> , <ref name="Reverse">PMID:10832092</ref> | ||
== Structural highlights == | == Structural highlights == | ||
CerN consists of two domains: a <scene name='91/910024/Domains/1'>catalytic domain near the N-terminal and an immunoglobulin-fold domain near the C-terminal</scene>.<ref name="Inoue">PMID:19088069</ref> Three β-sheets, each formed from four β-strands, compose a β-prism fold at the center of the N-terminal domain.<ref name="Inoue">PMID:19088069</ref> Surrounding the β-prism fold are 11 α-helices, forming an <scene name='91/910024/Bprism/3'>α+ β 2-layer sandwich fold</scene>.<ref name="Inoue">PMID:19088069</ref> The immunoglobulin-like C-terminal domain is composed of two β-sheets, containing four β-strands each, forming a <scene name='91/910024/Igfold/1'>β-sandwich fold</scene>.<ref name="Inoue">PMID:19088069</ref> Between the N- and C-terminal domains is a magnesium/calcium ion binding site that links together the two domains.<ref name="Inoue">PMID:19088069</ref> His37, Asp579, Asp581, and Thr854 interact with divalent cations within the <scene name='91/910024/Mg_bs/1'>magnesium/calcium ion binding site</scene>.<ref name="Inoue">PMID:19088069</ref> A <scene name='91/910024/Zinc_bs/3'>second metal-binding site containing a zinc ion</scene> is located within the N-terminal domain active site, where it is coordinated by His97, His204, Glu411, and a water molecule.<ref name="Inoue">PMID:19088069</ref> | CerN consists of two domains: a <scene name='91/910024/Domains/1'>catalytic domain near the N-terminal and an immunoglobulin-fold domain near the C-terminal</scene>.<ref name="Inoue">PMID:19088069</ref> Three β-sheets, each formed from four β-strands, compose a β-prism fold at the center of the N-terminal domain.<ref name="Inoue">PMID:19088069</ref> Surrounding the β-prism fold are 11 α-helices, forming an <scene name='91/910024/Bprism/3'>α+ β 2-layer sandwich fold</scene>.<ref name="Inoue">PMID:19088069</ref> The immunoglobulin-like C-terminal domain is composed of two β-sheets, containing four β-strands each, forming a <scene name='91/910024/Igfold/1'>β-sandwich fold</scene>.<ref name="Inoue">PMID:19088069</ref> Between the N- and C-terminal domains is a magnesium/calcium ion binding site that links together the two domains.<ref name="Inoue">PMID:19088069</ref> His37, Asp579, Asp581, and Thr854 interact with divalent cations within the <scene name='91/910024/Mg_bs/1'>magnesium/calcium ion binding site</scene>.<ref name="Inoue">PMID:19088069</ref> A <scene name='91/910024/Zinc_bs/3'>second metal-binding site containing a zinc ion</scene> is located within the N-terminal domain active site, where it is coordinated by His97, His204, Glu411, and a water molecule.<ref name="Inoue">PMID:19088069</ref> | ||
| Line 24: | Line 22: | ||
| - | == | + | == Evolutionary Conservation == |
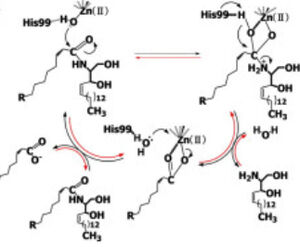
| - | + | Ceramidases are classified into three groups based upon primary structure and the optimal pH for their catabolic activity: acidic, neutral, and alkaline.<ref name="Ito">PMID:24064302</ref> '''Neutral ceramidases''' function optimally between pH 6.5-8.5 and can be found in eukaryotes and prokaryotes, while acidic and alkaline ceramidases are restricted to eukaryotes.<ref name="Ito">PMID:24064302</ref> The primary structure of neutral ceramidases is conserved from bacteria to humans, neutral ceramidase from ''Pseudomonas aeruginosa'' and humans share 32% identity and 47% similarity.<ref name="Ito">PMID:24064302</ref> The <scene name='91/910024/Activesite2/3'>active site</scene> amino acids responsible for the catalytic activity of neutral ceramidases, primarily His99 and Arg160, are conserved in ''P. aeruginosa'', humans, rats, fruit flies, and zebrafish, suggesting that the enzymatic mechanism is also shared ('''Figure 2''').<ref name="Okino">PMID:9603946</ref><ref name="Ito">PMID:24064302</ref> Furthermore, the crystal structure of Human neutral ceramidase ('''Figure 3''') is similar to that of P. aeruginosa, aligning with an RMSD value of 0.890 Å ('''Figure 4'''). However, the location of neutral ceramidase expression differs between bacteria/invertebrates and vertebrates.<ref name="Ito">PMID:24064302</ref> Neutral ceramidases from bacteria, slime molds, and fruit flies are secreted proteins, while the enzyme is primarily membrane-bound in vertebrates.<ref name="Ito">PMID:24064302</ref> Vertebrate ceramidases contain a serine/threonine/proline-rich domain, or mucin box, within the N-terminal region which is necessary for anchoring to the plasma membrane.<ref name="Ito">PMID:24064302</ref> | |
| + | |||
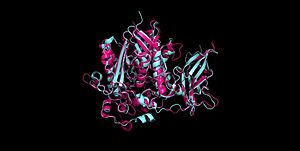
| + | [[Image:Aligned4wgk2zws.jpg|300px|right|thumb|'''Figure 4''' Alignment of Human neutral ceramidase [[4wgk]] (pink) and P. aeruginosa neutral ceramidase [[2zws]] (blue) RMSD=0.890 Å <ref name="Inoue">PMID:19088069</ref>]] | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 20:58, 2 May 2022
| |||||||||||