We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Vibriophage phiVC8 DpoZ
From Proteopedia
(Difference between revisions)
m |
m |
||
| Line 1: | Line 1: | ||
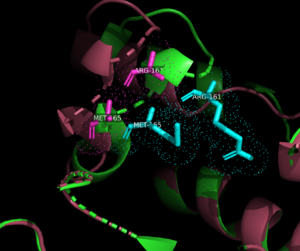
<StructureSection load='7pbk' size='340' side='right' caption='Vibriophage ΦVC8 DNA polymerase DpoZ deposited under the PDB ID [https://www.rcsb.org/structure/7PBK 7pbk]; thumb-exo open conformation.' scene='90/909993/Open_domains_colored/1'> | <StructureSection load='7pbk' size='340' side='right' caption='Vibriophage ΦVC8 DNA polymerase DpoZ deposited under the PDB ID [https://www.rcsb.org/structure/7PBK 7pbk]; thumb-exo open conformation.' scene='90/909993/Open_domains_colored/1'> | ||
== Introduction == | == Introduction == | ||
| - | [[Vibriophage phiVC8 DpoZ]] is a [[DNA polymerase]] belonging to the PolA family and the ΦVC8-like DpoZ subfamily, a group currently identified in certain species of bacteriophages<ref>PMID:34751404</ref>. DpoZ consists of two subfamilies: ΦVC8-like and Wayne-like. These polymerases confer selectivity in addition of the nucleobase 2-aminoadenine (Z) over adenine (A), with A completely ablated from their genomes. Z forms a non Watson-Crick base pair with thymine (T) consisting of three hydrogen bonds as opposed to the two present in A-T base pairing. | + | [[Vibriophage phiVC8 DpoZ]] is a [[DNA polymerase]] belonging to the PolA family and the ΦVC8-like DpoZ subfamily, a group currently identified in certain species of bacteriophages<ref>PMID:34751404</ref>. DpoZ consists of two subfamilies: ΦVC8-like and Wayne-like. These polymerases confer selectivity in addition of the nucleobase 2-aminoadenine (Z) over adenine (A), with A completely ablated from their genomes. Z forms a non Watson-Crick base pair with thymine (T) consisting of three hydrogen bonds as opposed to the two present in A-T base pairing. DpoZ is a relatively novel discovery, having only recently had its biosynthetic pathway described in detail<ref>PMID:33926954</ref>. DNA modifications in bacteriophages usually confer selective advantages by allowing phages to avoid host cell restriction enzyme digestion of their genomes<ref>Weigele, P., & Raleigh, E. A. (2016). Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. <i>Chemical Reviews</i>, <i>116</i>(20), 12655–12687. https://doi.org/10.1021/acs.chemrev.6b00114</ref>. The phage S-2L has a Z genome and encodes a [https://www.ncbi.nlm.nih.gov/gene/201973 PrimPol polymerase] as well as a nucleotide phosphohydrolase called [https://www.rcsb.org/structure/6ZPB DatZ] that removes the triphosphate tail of only dATP, excluding it from the genome<ref>PMID:33893297</ref>. Vibriophage ΦVC8 also encodes DatZ<ref>PMID:34751404</ref>, though it is unknown if it is the primary enzyme responsible for A exclusion or if its polymerase also plays a role. The specific mechanisms by which ΦVC8-like DpoZ polymerases carry out A exclusion and Z inclusion are still under investigation, though specific structural features and putative specificity mechanisms are highlighted below. |
== Structural Highlights == | == Structural Highlights == | ||
Revision as of 13:11, 3 May 2022
| |||||||||||
References
- ↑ Czernecki D, Hu H, Romoli F, Delarue M. Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage varphiVC8. Nucleic Acids Res. 2021 Nov 18;49(20):11974-11985. doi: 10.1093/nar/gkab955. PMID:34751404 doi:http://dx.doi.org/10.1093/nar/gkab955
- ↑ Zhou Y, Xu X, Wei Y, Cheng Y, Guo Y, Khudyakov I, Liu F, He P, Song Z, Li Z, Gao Y, Ang EL, Zhao H, Zhang Y, Zhao S. A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science. 2021 Apr 30;372(6541):512-516. doi: 10.1126/science.abe4882. PMID:33926954 doi:http://dx.doi.org/10.1126/science.abe4882
- ↑ Weigele, P., & Raleigh, E. A. (2016). Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. Chemical Reviews, 116(20), 12655–12687. https://doi.org/10.1021/acs.chemrev.6b00114
- ↑ Czernecki D, Legrand P, Tekpinar M, Rosario S, Kaminski PA, Delarue M. How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA. Nat Commun. 2021 Apr 23;12(1):2420. doi: 10.1038/s41467-021-22626-x. PMID:33893297 doi:http://dx.doi.org/10.1038/s41467-021-22626-x
- ↑ Czernecki D, Hu H, Romoli F, Delarue M. Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage varphiVC8. Nucleic Acids Res. 2021 Nov 18;49(20):11974-11985. doi: 10.1093/nar/gkab955. PMID:34751404 doi:http://dx.doi.org/10.1093/nar/gkab955
- ↑ Czernecki D, Hu H, Romoli F, Delarue M. Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage varphiVC8. Nucleic Acids Res. 2021 Nov 18;49(20):11974-11985. doi: 10.1093/nar/gkab955. PMID:34751404 doi:http://dx.doi.org/10.1093/nar/gkab955
- ↑ Czernecki D, Hu H, Romoli F, Delarue M. Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage varphiVC8. Nucleic Acids Res. 2021 Nov 18;49(20):11974-11985. doi: 10.1093/nar/gkab955. PMID:34751404 doi:http://dx.doi.org/10.1093/nar/gkab955
- ↑ Miller, B.R., Beese,L.S., Parish, C.A. and Wu,E.Y. (2015) The closing mechanism of DNA polymerase I at atomic resolution. Structure, 23,1609–1620. https://doi.org/10.1016/j.str.2015.06.016
- ↑ Czernecki D, Hu H, Romoli F, Delarue M. Structural dynamics and determinants of 2-aminoadenine specificity in DNA polymerase DpoZ of vibriophage varphiVC8. Nucleic Acids Res. 2021 Nov 18;49(20):11974-11985. doi: 10.1093/nar/gkab955. PMID:34751404 doi:http://dx.doi.org/10.1093/nar/gkab955
- ↑ Tabor, S., & Richardson, C. C. (1995). A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proceedings of the National Academy of Sciences of the United States of America, 92(14), 6339–6343. https://doi.org/10.1073/pnas.92.14.6339
- ↑ Suzuki, M., Baskin, D., Hood, L., & Loeb, L. A. (1996). Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proceedings of the National Academy of Sciences of the United States of America, 93(18), 9670–9675. https://doi.org/10.1073/pnas.93.18.9670
- ↑ Juarez-Quintero, V., Peralta-Castro, A., Benítez Cardoza, C. G., Ellenberger, T. & Brieba, L. G. (2021). Structure of an open conformation of T7 DNA polymerase reveals novel structural features regulating primer-template stabilization at the polymerization active site. Biochemical Journal, 478, 2665–2679https://doi.org/10.1042/BCJ20200922
- ↑ Samson, C., Legrand,P., Tekpinar,M., Rozenski,J., Abramov,M., Holliger,P., Pinheiro,V.B., Herdewijn, P. and Delarue,M. (2020) Structural studies of HNA substrate specificity in mutants of an archaeal DNA polymerase obtained by directed evolution. Biomolecules, 10, 1647.