Structural Definition of Polyspecific Compensatory Ligand Recognition by P-glycoprotein
Christina A. Le, Daniel S. Harvey and Stephen G. Aller [1]
Molecular Tour
Most cell types including healthy- and cancerous- cells deal with threats from chemicals and toxins by a number of different mechanisms that allow them to develop resistance to toxic side-effects. A major mechanism includes the actual pumping of the threatening molecules out of the cell before they can pass all the way into the cell to cause damage. In the case of cancer chemotherapies, FDA-approved drugs act as the toxins to cancer cells, but the cancer cells can also adapt by increasing the number of molecular pumps at the membranous cell surface to deal with the threat and acquire resistance. One of the most dominant molecular pumps in the human body is called P-glycoprotein (MDR1/ABCB1) or Pgp for short. Also referred to as the multidrug resistance transporter, Pgp is capable of recognizing and expelling perhaps thousands of small molecule drugs and toxins to protect cells. An early idea involved developing a second drug that might 'inhibit' Pgp and allow the original chemotherapeutic drug to penetrate the cell and become more effective again. The most potent Pgp inhibitors developed, however, appeared to inhibit Pgp systemically, i.e. at important locations such as the liver, where the continual function of Pgp in detoxification is critical. Such inhibitors resulted in worse toxic side effects and even fatality. A more recent idea is to carefully design/modify the chemotherapeutics themselves such that they might not be efficiently recognized by Pgp and might evade transport. The latter idea is certainly very daunting given the polyspecific nature of Pgp - in other words, the fact that the Pgp transporter seems to have evolved to be able to recognize and transport a very large number of chemically different molecules suggests that the design of evaders will be very challenging.
Of high scientific value would be a better understanding of the molecular mechanisms of Pgp that confer its polyspecific abilities. A major part of this quest by several groups has been the determination of Pgp structure to near-atomic precision, including in the presence of ligands (drugs and drug-like molecules that are recognized and bound to the Pgp protein). The Pgp pump is now known to rock back-and-forth from an inverted "V"-shape to a "V"-shape conformation during its pumping activity. Pgp uses (hydrolyzes) ATP as an energy source to power the pumping process as it changes protein conformations. The binding location of several ligands is known to shift depending on even slight changes in protein conformation, hinting at a mechanism of polyspecificity. Furthermore, distinct ligand binding sites have been described for certain ligands whereas some ligands share overlapping sites with other ligands. What was not known until now is that, if the protein conformation of Pgp was fixed in place, but the normal ligand binding site was slightly perturbed (i.e. by mutagenesis of a single ligand-interacting amino acid residue), would the ligand stay in the same site or would it shift to new sites?
In our work here, we used an environmental toxin, called BDE-100, that is known to bind to Pgp in previous work by a highly precise and accurate technique called x-ray crystallography (XRC). We reproduced the exact conformation of Pgp including the precise location of BDE-100 binding. Next, we mutated each of five single amino acids that were in direct contact with, or very close to, the BDE-100 ligand. Of particular importance, most of the amino acid residues that form ligand binding sites are of a specific class called aromatic residues such as tyrosine (Y) and phenylalanine (F). We mutated five aromatic residues, one at a time, that interacted with BDE-100, to a much smaller amino acid (alanine, A) that is not aromatic in character (F979A, Y303A, Y306A, F724A and F728A), and determined if BDE-100 stayed in the same place or moved to another location. BDE-100 also has strong advantages because it contains bromine atoms which are easy to localize by XRC using a process called anomalous diffraction. Two mutants, Y303A and Y306A showed strong BDE-100 occupancy at the original ligand binding site (we call site 1), but also revealed a novel site 2 located on the opposing pseudo-symmetric half of the drug binding pocket (DBP). Surprisingly, the F724A structure of Pgp had no detectable binding in site 1 but exhibited a novel site shifted 11 Å from site 1. ATPase studies revealed shifts in ATPase kinetics for the five mutants but otherwise indicated catalytically active transporter that was inhibited by BDE-100 similar to unmutated (wildtype) Pgp. Our results emphasize a high degree of compensatory drug recognition in Pgp made possible by aromatic amino acid sidechains concentrated in the DBP. Compensatory recognition forms the underpinning of polyspecific drug transport but also highlights the challenges associated with the design of therapeutics that evade efflux altogether.
Six Pgp mutants and localization of BDE-100 by X-ray crystallography:
Views are from the extracellular side perpendicular to the membrane. BDE-100 in each structure is shown in stick representation with Br atoms colored crimson. Phe974, which undergoes a disorder-to-order transition upon binding in site 2, is labeled in all six scenes.
Superposition of BDE-100 for all six mutants (C952A parent, F979A, F724A, F728A, Y303A and Y306A) shown for the inward-facing conformation of Pgp, depicting the relative localization within the DBP:
The ligand in C952A is indistinguishable from that in F979A. The ligand in C952A is colored dark blue, that in F724A is colored yellow, that in Y303A is colored light green, that in Y306A is colored dark green and that in F728A is colored orange.
Close-ups of the alternate BDE-100 binding sites:
Aromatic side chains in the DBP that play a role in binding in at least one site (=< 4.2 Å from the ligand) are shown as sticks colored tan and labeled.
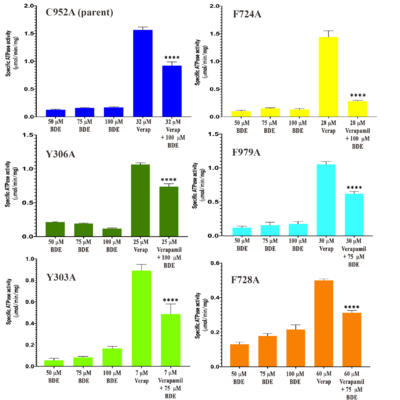
Figure 3. The effect of BDE-100 on the verapamil-stimulated ATPase activity of the six Pgp mutations. The effect of BDE-100 in the absence of verapamil on each mutant was determined at 50, 75 and 100 μM. The EC
50 of verapamil-stimulated ATPase activity of each mutant was pre-determined (Supplementary Fig. S4) and is shown here. The level of BDE-100 inhibition of the verapamil-stimulated ATPase EC
50 for each mutant is also shown. P values were were determined from a two-tailed unequal variance test: ****,
p =< 0.0001; ***,
p =< 0.0001–0.001; *,
p =< 0.05–0.01. The scale of each graph is represented to enhance the differences of verapamil concentration on ATPase for the given mutant, particularly since one mutant, F728A, exhibited a considerably lower
Vmax compared with the other mutants.
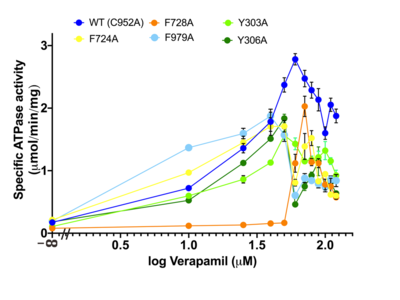
Figure 4. Dose–response curve of verapamil-stimulated ATPase activity. Curves for each different mutant are colored according to the legend of the figure. The F728A mutant exhibited the highest EC
50 of 60 μM, followed by C952A (32 μM), F979A (30 μM), F724A (28 μM), Y306A (25 μM) and Y303A (7.3 μM). All measurements were determined with seven independent measurements (
n = 7).
. Transmembrane (TM) α-helices are are colored from the N-terminus to the C-terminus (blue to red). Two critical TM helices known to bind multiple different ligands, TM6 and TM12, are labeled. Dual DBE-100 occupancy for the Y303A, Y306A and F728A mutants was apparent, in which the two binding sites are in opposing ‘halves’ of the pseudosymmetric DBP. The F724A mutant structure was distinct from all others in that it contains a single BDE-100 bound in a novel site that lies on the axis of pseudosymmetry (dotted line), which we designate site 3. The rationale for the site nomenclature is as follows. Site 1 is the canonical site previously discovered by Nicklisch et al. (2016)[2]. Some structures with site 1 or site 1A occupancy also had a second site occupancy, which we designate site 2. Site 3 is the distinct novel single-site binding unique to F724A.
PDB references: P-glycoprotein, C952A mutant, complex with BDE-100, 6unj; C952A/F979A mutant, complex with BDE-100, 6ujp; C952A/F724A mutant, complex with BDE-100, 6ujr; C952A/F728A mutant, complex with BDE-100, 6ujs; C952A/Y303A mutant, complex with BDE-100, 6ujt; C952A/Y306A mutant, complex with BDE-100, 6ujw.
References
- ↑ Le CA, Harvey DS, Aller SG. Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein. IUCrJ. 2020 Jun 6;7(Pt 4):663-672. doi: 10.1107/S2052252520005709. eCollection, 2020 Jul 1. PMID:32695413 doi:http://dx.doi.org/10.1107/S2052252520005709
- ↑ Nicklisch SC, Rees SD, McGrath AP, Gokirmak T, Bonito LT, Vermeer LM, Cregger C, Loewen G, Sandin S, Chang G, Hamdoun A. Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure. Sci Adv. 2016 Apr 15;2(4):e1600001. doi: 10.1126/sciadv.1600001. eCollection 2016, Apr. PMID:27152359 doi:http://dx.doi.org/10.1126/sciadv.1600001




