This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Art:Dodecapod
From Proteopedia
| Line 4: | Line 4: | ||
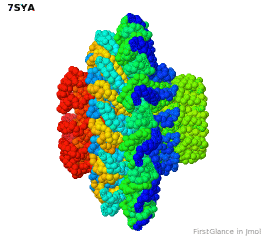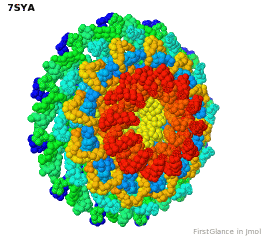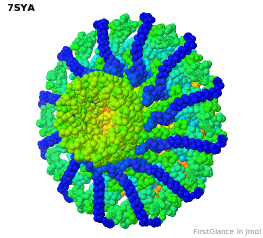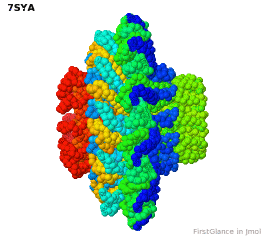
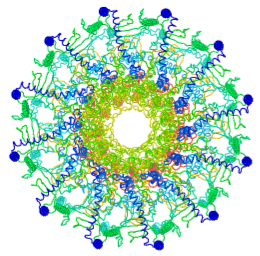
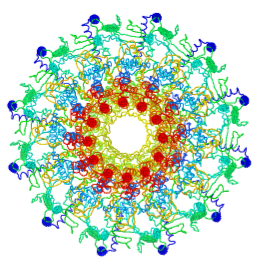
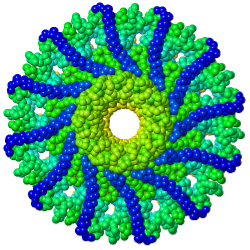
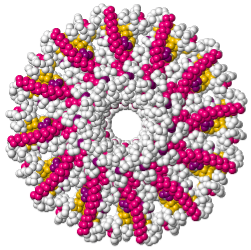
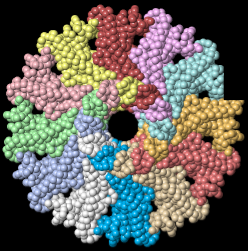
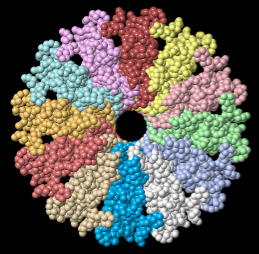
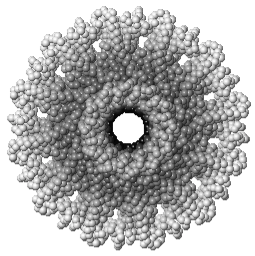
Tailed bacteriophages and herpesviruses have a DNA "pipe" in their capsids, called a portal protein<ref name="hou2022">PMID: 35278476</ref>. The images here depict [[7sya]], a homo-dodecamer<ref name="hou2022" />. In the animation at right, each of the 12 protein chains is colored amino to carboxy spectrum (N->C Rainbow): | Tailed bacteriophages and herpesviruses have a DNA "pipe" in their capsids, called a portal protein<ref name="hou2022">PMID: 35278476</ref>. The images here depict [[7sya]], a homo-dodecamer<ref name="hou2022" />. In the animation at right, each of the 12 protein chains is colored amino to carboxy spectrum (N->C Rainbow): | ||
{{Template:ColorKey_N2CRainbow}} | {{Template:ColorKey_N2CRainbow}} | ||
| - | All images and the animation were generated with [[FirstGlance in Jmol]] (version 4.0, not yet released in mid-May 2022), most using color schemes available under ''Solid'' (Views tab). The two faces of the oblate structure (or tube with a flange) are designated the N-face (face with amino termini) and the C-face (face with carboxy termini). | + | All images show a model simplified to alpha carbons only, with the alpha carbons rendered at an exaggerated diameter (6.0 Å) to give a solid appearance. All images and the animation were generated with [[FirstGlance in Jmol]] (version 4.0, not yet released in mid-May 2022), most using color schemes available under ''Solid'' (Views tab). The two faces of the oblate structure (or tube with a flange) are designated the N-face (face with amino termini) and the C-face (face with carboxy termini). |
<table class="wikitable"> | <table class="wikitable"> | ||
Revision as of 15:03, 17 May 2022
Tailed bacteriophages and herpesviruses have a DNA "pipe" in their capsids, called a portal protein[1]. The images here depict 7sya, a homo-dodecamer[1]. In the animation at right, each of the 12 protein chains is colored amino to carboxy spectrum (N->C Rainbow):
| N | C |
All images show a model simplified to alpha carbons only, with the alpha carbons rendered at an exaggerated diameter (6.0 Å) to give a solid appearance. All images and the animation were generated with FirstGlance in Jmol (version 4.0, not yet released in mid-May 2022), most using color schemes available under Solid (Views tab). The two faces of the oblate structure (or tube with a flange) are designated the N-face (face with amino termini) and the C-face (face with carboxy termini).
|
|
|
|
|
|
References & Notes
- ↑ 1.0 1.1 David Hou CF, Swanson NA, Li F, Yang R, Lokareddy RK, Cingolani G. Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3. J Mol Biol. 2022 Mar 9;434(9):167537. doi: 10.1016/j.jmb.2022.167537. PMID:35278476 doi:http://dx.doi.org/10.1016/j.jmb.2022.167537