This is a default text for your page Max Hideki Oliveira Homma/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
Ribonucleotide reductase (RNR) is an enzyme responsible for converting deoxyribonucleotides (dNTPs) from ribonucleotides (NTPs). It catalyzes this chemical reaction by removing the 2'-hydroxyl group of ribose ring of ribonucleotides. Therefore, this enzyme is also known as ribonucleoside diphosphate reductase (rNDP). In this way, this enzyme promotes the synthesis of precursors for regeneration and construction of DNA.
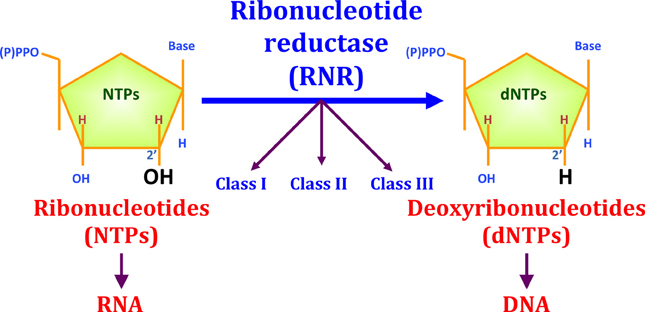
Because of its essential function for the structure of DNA, knowing that all cellular organisms have DNA as a hereditary structure, it is likely that RNR is present in all growing cells of all living beings. In addition, it is also speculated that the RNR played a key role in the transition from the "RNA world" to the "DNA world". In this question, RNR are subdivided into three classes, and the third class can function without oxygen and uses simple structures, thus, it is believed that this class is the oldest.
Classification and distribuition
RNRs can be classified into three distinct classes according to the method of generating thiyl ions triggered by the presence of a particular cofactor. RNRs I are characterized by having a complex quaternary structure and dependence on dioxygen to assemble the cysteine oxidant. RNRs I are present in viruses and in all domains of life and can be subdivided into RNR Ia, Ib, Ic, Id and Ie. RNRs Ia are the best characterized and will receive greater focus on this page. These enzymes present the diiron-tyrosyl radical and are composed of two types of subunits called R1 (or alpha) and R2 (or beta). The biggest difference between the Ia RNRs and the other class I RNRs is in the substitution of diiron-tyrosyl for other cofactors. For example, in RNR Ib, the two ferric ions are replaced by manganese. The cofactors present in the different class I RNRs are shown in the figure.
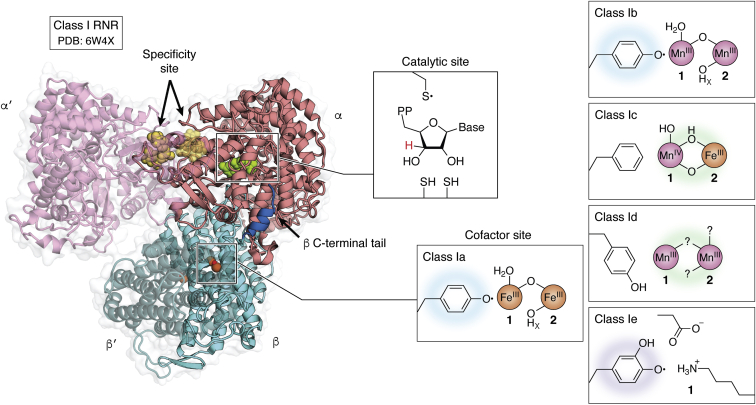
Although less studied, there are also class II and III RNRs, which are found only in microorganisms. Class II RNR has adenosylcobalamin, a vitamin B12 derivative, as a cofactor. Class III RNRs, on the other hand, use a [4Fe-4S]-activase to generate a stable glycyl radical capable of leading to the generation of the oxidant Cys at the active site of the RNR. The presence of RNRs of classes II and III is usually related to a better adaptation in environments with low oxygen availability, and RNRs III usually have their activity inhibited by oxygen. Complex eukaryotes, as they generally encode only RNR Ia, have low occupancy in hypoxic environments. Although they share an almost universal ribonucleotide reduction mechanism, the three classes of RNRs have low primary sequence similarity to each other, which may suggest independent emergence.
Several microorganisms are able to encode RNRs of different classes. In the case of Escherichia coli, RNR Ia is preferentially expressed in the growth phase under aerobic conditions, while RNR III is mainly expressed under anaerobic conditions. Furthermore, in environments with low iron availability or when there is biofilm formation, E. coli tends to preferentially express RNR Ib.
Geral structure of RNR Ia
Among the types of RNR, Ia is the best characterized, grouped among the class I RNRs, enzymes Oxygen dependent and found in all eukaryotic organisms and in some prokaryotes and viruses.
The RNR class Ia is a structure that has the diiron-tyrosyl radical and is composed of two subunits, R1 (or alpha) and R2 (or beta). The large R1 structure, also known as alpha2-homodimer, encoded by the NrdA genes, contains the active site for nucleotide binding and reduction and two allosteric sites, involved in the allosteric regulation of substrate specificity and general enzyme activity.
The small R2 structure, also known as beta2-homodimer, encoded by the Nrdb genes, contains a binding site for two iron ions in each polypeptide chain, where the interaction with the metallic cofactor occurs. Metal ion transitions are necessary cofactors to reduce nucleotides, as they generate an oxidant to an active cysteine site (Cys), producing a thiyl radical that is necessary for the initiation of catalysis in all RNRs.
Structural highlights
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.


