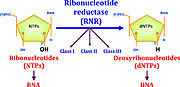
User:Max Hideki Oliveira Homma/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
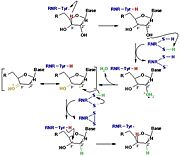
[[Image:RNR3.jpg|thumb|'''Figure 3:''' The reduction of ribonucleotides catalyzed by the RNR involves the participation of cysteines present in the active site]] | [[Image:RNR3.jpg|thumb|'''Figure 3:''' The reduction of ribonucleotides catalyzed by the RNR involves the participation of cysteines present in the active site]] | ||
| - | The catalysis mechanism performed at the RNR active site is represented in the figure 3 and depends of <scene name='91/910665/Cys_of_active_site/1'> | + | The catalysis mechanism performed at the RNR active site is represented in the figure 3 and depends of <scene name='91/910665/Cys_of_active_site/1'>cysteines of active site</scene>.Initially, the hydrogen bonded to the 3' carbon of the ribose ribonucleotide is transferred to the thiyl radical of Cys439. As a result, the 2'OH radical becomes more accessible for acid hydrolysis and its protonation is catalyzed by Cys225 and Glu441, resulting in the exit of a molecule from the substrate. After that, a proton is transferred from Cys462 to Cys225 which then transfers a hydrogen to the 2' carbon. Then, the transfer of a hydrogen to the 2' carbon of the substrate occurs concomitantly with the formation of a <scene name='91/910665/Cys_ball_and_stick/1'>disulfide bridge</scene> between the Cys225 and Cys462 of the RNR. Finally, till radical regeneration occurs with the transfer of a hydrogen from Cys439 to the 3' carbon of the substrate, leading to the release of a deoxyribonucleotide. At the end of the catalysis, the disulfide bridge between Cys225 and Cys462 must still be reduced. The electron used in this reduction comes from a redox chain whose initial donor is NADPH. Numerous components participate in this redox chain, including Cys 754 and Cys759 from R1 itself. The reduction of the disulfide bridge between Cys225 and Cys462 would be very difficult to occur if there was a radical very close to the disulfide bridge, since the tendency would be for this radical to be reduced instead of the disulfide bridge. This is a likely explanation for the existence of a second subunit in the RNR, as the R2 subunit harbors a tyrosyl radical that is difficult to reduce. |
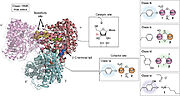
== R2 subunit and cofactor == | == R2 subunit and cofactor == | ||
| Line 49: | Line 49: | ||
== Regulation == | == Regulation == | ||
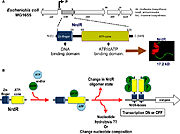
| - | The regulation of RNRs is very refined and occurs at several levels. In microorganisms, a transcription factor, called NrdR, was identified, which is capable of binding to NrdR-box, a consensus sequence present in the promoters of nrd genes, which encode RNRs. The NrdR is composed of two domains, the Zn-finger, which binds to DNA, and the ATP-cone, which has ATP and/or dATP binding sites. Although its regulatory mechanism has not been fully elucidated, it is proposed that the binding of dATP to the ATP-cone causes NrdR to dimerize and interact more with the NrdR-boxes sequences, leading to reduced transcription of nrd genes. | + | The regulation of RNRs is very refined and occurs at several levels. In microorganisms, a transcription factor, called NrdR, was identified, which is capable of binding to NrdR-box, a consensus sequence present in the promoters of nrd genes, which encode RNRs. The NrdR is composed of two domains, the Zn-finger, which binds to DNA, and the ATP-cone, which has ATP and/or dATP binding sites. Although its regulatory mechanism has not been fully elucidated, it is proposed (figure 5) that the binding of dATP to the ATP-cone causes NrdR to dimerize and interact more with the NrdR-boxes sequences, leading to reduced transcription of nrd genes. |
[[Image:RNR5.jpg|thumb|'''Figure 5:''' proposed model of how NrdR modulates the expression of nrd genes in bacteria (Torrents, 2014)]] | [[Image:RNR5.jpg|thumb|'''Figure 5:''' proposed model of how NrdR modulates the expression of nrd genes in bacteria (Torrents, 2014)]] | ||
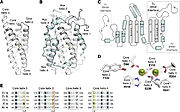
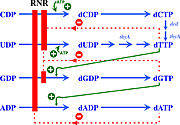
| - | The activity of the already translated RNR is also finely regulated. Although RNRs are capable of reducing the 4 ribonucleotides, the binding of dATP (or ATP), dGTP, dCTP and dUTP to the S site of the R1 subunit changes which ribonucleotide will be preferentially reduced by the enzyme. For example, in Ia RNRs, the binding of dATP or ATP to the S site causes the enzyme to reduce CDP and UDP while inhibiting the reduction of ADP. | + | The activity of the already translated RNR is also finely regulated. Although RNRs are capable of reducing the 4 ribonucleotides, the binding of dATP (or ATP), dGTP, dCTP and dUTP to the S site of the R1 subunit changes which ribonucleotide will be preferentially reduced by the enzyme. For example, in Ia RNRs, the binding of dATP or ATP to the S site causes the enzyme to reduce CDP and UDP while inhibiting the reduction of ADP (figure 6). |
[[Image:RNR6.jpg|thumb|'''Figure 6:''' Modulation of RNR Ia activity by binding deoxyribonucleotide triphosphates to the S site (Torrents, 2014).]] | [[Image:RNR6.jpg|thumb|'''Figure 6:''' Modulation of RNR Ia activity by binding deoxyribonucleotide triphosphates to the S site (Torrents, 2014).]] | ||
| - | In addition to the S site, the A site is also capable of regulating RNR Ia activity. In this sense, ATP binding to this site will stimulate the general activity of the enzyme, | + | In addition to the S site, the A site is also capable of regulating RNR Ia activity. In this sense, ATP binding to this site will stimulate the general activity of the enzyme. On the other hand, the binding of <scene name='91/910665/R1_inhibition/1'>dATP to the A site</scene> of the R1 subunit inhibits the activity of RNR Ia by altering its oligomerization state, forming a <scene name='91/910665/Rnr_ia_inhibited/1'>complex</scene> composed of 2 R1 subunits and two R2 subunits. However, because dATP binds with a much higher affinity to the S site than to the A site, its inhibitory effect is only significant when this molecule is in high concentrations. |
Due to the cellular importance of RNRs and the possibility of regulating their activity, inhibitors of these enzymes have been used for therapeutic purposes. Inhibition of Ia RNRs seems to be a particularly interesting strategy in the fight against cancer, as it leads to the interruption of cell proliferation. An alternative R2 subunit, called p53R2, was also identified in mammalian cells, which is associated with the DNA repair system and is often not functioning correctly in cancer cells. | Due to the cellular importance of RNRs and the possibility of regulating their activity, inhibitors of these enzymes have been used for therapeutic purposes. Inhibition of Ia RNRs seems to be a particularly interesting strategy in the fight against cancer, as it leads to the interruption of cell proliferation. An alternative R2 subunit, called p53R2, was also identified in mammalian cells, which is associated with the DNA repair system and is often not functioning correctly in cancer cells. | ||
Inhibitors of Ia RNRs can be divided into three main groups. The first group consists of inhibitors that bind to the R1 subunit, either at the active site or at allosteric sites. The second group comprises inhibitors that act on the R2 subunit, usually being iron-chelating or radical-scavenging compounds. Finally, the third group is composed of inhibitors that form hairpins with the mRNA transcribed from the nrd genes, preventing their translation. In addition to these three groups, there are also inhibitors that prevent the union between the R1 and R2 subunits. | Inhibitors of Ia RNRs can be divided into three main groups. The first group consists of inhibitors that bind to the R1 subunit, either at the active site or at allosteric sites. The second group comprises inhibitors that act on the R2 subunit, usually being iron-chelating or radical-scavenging compounds. Finally, the third group is composed of inhibitors that form hairpins with the mRNA transcribed from the nrd genes, preventing their translation. In addition to these three groups, there are also inhibitors that prevent the union between the R1 and R2 subunits. | ||
| - | |||
| - | |||
| - | <scene name='91/910665/Rnr_ia_inhibited/1'>Inhibition of RNR Ia by dATP</scene> | ||
| - | |||
| - | <scene name='91/910665/R1_inhibition/1'>R1 inhibition by dATP and dADP</scene> | ||
Revision as of 19:51, 16 June 2022
</math>==Your Heading Here (maybe something like 'Structure')==
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644