User:Sabrina K.K. Komatsu/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Structure=== | ===Structure=== | ||
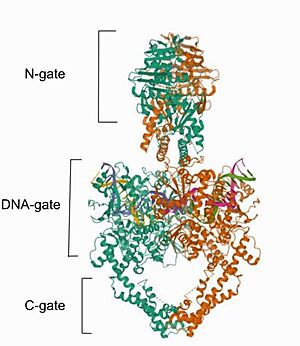
| - | Cryo-EM reconstructions of the hTop2α nucleoprotein complex showed that Mammalian <scene name='91/914441/Topoisomerase_ii_alfa/1'>DNA topoisomerase IIα (human) IIA</scene> is a homodimeric protein, in whitch each subunit structure can be broken down into three major components that are connected by hinged like regions: the N gate, that contains the ATPase domain, the DNA gate and the C gate. (Broek et al, 2020). Biochemical and structural studies have shown that the ATPase domain or N-gate, and the DNA binding/cleavage domain forming the DNA- and C-gates, are allosterically connected, a key feature of its activity (Broek et al, 2020). Furthermore, the literature contains various descriptions of the structures of the N-gate and DNA-gate, but the C-gate is less explored. [[Image: | + | Cryo-EM reconstructions of the hTop2α nucleoprotein complex showed that Mammalian <scene name='91/914441/Topoisomerase_ii_alfa/1'>DNA topoisomerase IIα (human) IIA</scene> is a homodimeric protein, in whitch each subunit structure can be broken down into three major components that are connected by hinged like regions: the N gate, that contains the ATPase domain, the DNA gate and the C gate. (Broek et al, 2020). Biochemical and structural studies have shown that the ATPase domain or N-gate, and the DNA binding/cleavage domain forming the DNA- and C-gates, are allosterically connected, a key feature of its activity (Broek et al, 2020). Furthermore, the literature contains various descriptions of the structures of the N-gate and DNA-gate, but the C-gate is less explored. [[Image:Image- Top2.jpg|300px|right|thumb|Image from the RCSB PDB (https://www.rcsb.org/structure/6ZY8) of PDB ID 6ZY8, Cryo-EM structure of the entire Human topoisomerase II alpha in State 2.]] |
| + | The protein DNA Topoisomerase IIα (human) has two <scene name='91/914441/Chains/2'>chains</scene> one represented in <scene name='91/914441/Green/1'>green</scene> and one in <scene name='91/914441/Gray/1'>gray</scene>, both bounded to <scene name='91/914441/Dna/1'>double-stranded DNA</scene>. | ||
| + | |||
| + | The <scene name='91/914441/Secondary/1'>secondary structure</scene> of this protein has <scene name='91/914441/Sheet/1'>beta sheets</scene> and <scene name='91/914441/Alpha_helice/1'>alpha helices</scene>. <scene name='91/914441/Secondary_monomer/1'>Here</scene> is what a monomer secondary structure looks like. In other representation, the <scene name='91/914441/C_to_n/2'>C-terminus and the N-terminus</scene> can be seen in red and dark blue respectively. | ||
| - | ==The N gate== | + | ===The N gate=== |
The N-gate contains an ATPase domain, which was resolved to 1.86 Å by diffraction data of crystals, acquired using synchrotron radiation. The ATPase domain structure contains one dimer per asymmetric unit (Wei et al, 2005). The polypeptide chain folds into two discrete structural modules, with the N-terminal module bearing a similarity with the GHKL superfamily, consisting of an eight-stranded, mixed-sheet backed by four -helices. The C-terminal module structure consists of a four-stranded, mixed -sheet with one -helix on one face and two on the other, and has the role of communicating conformational information between the ATPase domain and the DNA gate. The primary dimerization interface involves the nucleotide-binding modules of the two monomers, with two short helices from the N-terminal of the two monomers interacting exclusively with the nucleotide-binding module of the C2 symmetry-related monomer. Furthermore, in each one of the monomers, residues of the nucleotide-binding module form one nucleotide-binding pocket. Loading of ATP onto the ATPase domain triggers dimerization and temporal capture of the DNA strands and that dimerization of the ATPase domain is nucleotide-dependent. Interestingly, the cavity in the domain is not big enough to hold the DNA segment, requesting an extension, but the study indicated that expansion is likely to occur by rigid-body displacement of the transducer module. This movement it is known to cause a retraction of the Lys-378 -amino group from the -phosphat, which is essential for ATPase activity (Schmidt, Osheroff & Berger, 2012) | The N-gate contains an ATPase domain, which was resolved to 1.86 Å by diffraction data of crystals, acquired using synchrotron radiation. The ATPase domain structure contains one dimer per asymmetric unit (Wei et al, 2005). The polypeptide chain folds into two discrete structural modules, with the N-terminal module bearing a similarity with the GHKL superfamily, consisting of an eight-stranded, mixed-sheet backed by four -helices. The C-terminal module structure consists of a four-stranded, mixed -sheet with one -helix on one face and two on the other, and has the role of communicating conformational information between the ATPase domain and the DNA gate. The primary dimerization interface involves the nucleotide-binding modules of the two monomers, with two short helices from the N-terminal of the two monomers interacting exclusively with the nucleotide-binding module of the C2 symmetry-related monomer. Furthermore, in each one of the monomers, residues of the nucleotide-binding module form one nucleotide-binding pocket. Loading of ATP onto the ATPase domain triggers dimerization and temporal capture of the DNA strands and that dimerization of the ATPase domain is nucleotide-dependent. Interestingly, the cavity in the domain is not big enough to hold the DNA segment, requesting an extension, but the study indicated that expansion is likely to occur by rigid-body displacement of the transducer module. This movement it is known to cause a retraction of the Lys-378 -amino group from the -phosphat, which is essential for ATPase activity (Schmidt, Osheroff & Berger, 2012) | ||
| - | ==The DNA gate== | + | ===The DNA gate=== |
Within the DNA gate there are three important domains: the topoisomerase primases (toprim domain), the winged helix domain (WHD domain or 5Y-CAP)) and the tower domain. The TOPRIM domain contains a DxD motif, where metal ion binding occurs due to two aspartates at positions 541 and 543, which coordinates a single magnesium 2 plus ion quelation,and a glutamate residue, that donates a proton to the sugar hydroxyl of the DNA during cleavage and abstracting the proton from the 3ʹ-OH during re-ligation. The TOPRIM domain also contributes to DNA binding, due to conserved residues, namely the EGDS and PLRGK motifs, which interact with the G-segment. Furthermore, the TOPRIM employs a distinct insertion, called insertion 2, composed of a short helix followed by a long loop, known for accommodation of DNA (Chang et al, 2013) | Within the DNA gate there are three important domains: the topoisomerase primases (toprim domain), the winged helix domain (WHD domain or 5Y-CAP)) and the tower domain. The TOPRIM domain contains a DxD motif, where metal ion binding occurs due to two aspartates at positions 541 and 543, which coordinates a single magnesium 2 plus ion quelation,and a glutamate residue, that donates a proton to the sugar hydroxyl of the DNA during cleavage and abstracting the proton from the 3ʹ-OH during re-ligation. The TOPRIM domain also contributes to DNA binding, due to conserved residues, namely the EGDS and PLRGK motifs, which interact with the G-segment. Furthermore, the TOPRIM employs a distinct insertion, called insertion 2, composed of a short helix followed by a long loop, known for accommodation of DNA (Chang et al, 2013) | ||
The WHD contains a helix-turn-helix fold, common in DNA-binding proteins, that have catalytic tyrosine residues in the C-terminal helix (also termed the ‘recognition helix’), responsible for forming a reversible covalent bond with the 5ʹ-scissile DNA phosphate. Besides that, The WHD also holds an isoleucine, which intercalates into the G-segment (the first segment of DNA duplex that enter the enzyme) producing a ∼150° bend, promoting DNA cleavage. The cleaving of the DNA backbone occurs in a bipartite active site formed by the TOPRIM DxD motif and the active site tyrosine of the WHD, which physically associate in different orientations during the enzyme's cycle of action (Chang et al, 2013) | The WHD contains a helix-turn-helix fold, common in DNA-binding proteins, that have catalytic tyrosine residues in the C-terminal helix (also termed the ‘recognition helix’), responsible for forming a reversible covalent bond with the 5ʹ-scissile DNA phosphate. Besides that, The WHD also holds an isoleucine, which intercalates into the G-segment (the first segment of DNA duplex that enter the enzyme) producing a ∼150° bend, promoting DNA cleavage. The cleaving of the DNA backbone occurs in a bipartite active site formed by the TOPRIM DxD motif and the active site tyrosine of the WHD, which physically associate in different orientations during the enzyme's cycle of action (Chang et al, 2013) | ||
| - | ==The tower domain== | + | ===The tower domain=== |
The tower domain (TD) is also involved with DNA binding. G-segment binds with a beta-sheet region, while another region composed by a + b sheets mediates domain-domain interactions (Chang et al, 2013).The TD DNA-binding surface juxtaposes with the DNA-intercalating wing-2 motif of WHD, extending the DNA-binding surface of the WHD and anchoring the outer end of the bent G-segment DNA. Interestingly, the structural conservation observed for the fixed packing between the tower domain and WHD suggests their assembly as an integral unit for the production and stabilization of DNA-bending (Chang et al, 2013). | The tower domain (TD) is also involved with DNA binding. G-segment binds with a beta-sheet region, while another region composed by a + b sheets mediates domain-domain interactions (Chang et al, 2013).The TD DNA-binding surface juxtaposes with the DNA-intercalating wing-2 motif of WHD, extending the DNA-binding surface of the WHD and anchoring the outer end of the bent G-segment DNA. Interestingly, the structural conservation observed for the fixed packing between the tower domain and WHD suggests their assembly as an integral unit for the production and stabilization of DNA-bending (Chang et al, 2013). | ||
| Line 46: | Line 49: | ||
| - | == Structural highlights == | ||
| - | The monomers secondary structures: <scene name='91/914441/Secondary_structure_1/1'>1</scene> | ||
| - | |||
| - | |||
| - | The protein DNA Topoisomerase IIα (human) has two <scene name='91/914441/Chains/2'>chains</scene> one represented in <scene name='91/914441/Green/1'>green</scene> and one in <scene name='91/914441/Gray/1'>gray</scene>, both bounded to <scene name='91/914441/Dna/1'>double-stranded DNA</scene>. | ||
| - | |||
| - | The <scene name='91/914441/Secondary/1'>secondary structure</scene> of this protein has <scene name='91/914441/Sheet/1'>beta sheets</scene> and <scene name='91/914441/Alpha_helice/1'>alpha helices</scene>. <scene name='91/914441/Secondary_monomer/1'>Here</scene> is what a monomer secondary structure looks like. In other representation, the <scene name='91/914441/C_to_n/2'>C-terminus and the N-terminus</scene> can be seen in red and dark blue respectively. | ||
| - | |||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 19:35, 18 June 2022
DNA Topoisomerase II
| |||||||||||