User:Isabela de Aquino Zogbi/Sandbox1
From Proteopedia
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
| - | <scene name='91/915204/Dysferlin/2'>Dysferlin</scene> is a large transmembrane protein of approximately 230kDa encoded by the dysferlin gene (DYSF omim) highly expressed in striated skeletal and cardiac muscle, but can be found in kidney, placenta, lung and brain tissues<ref[3]>. Dysferlin is a protein that belongs to the same family of genes as ''Caenorhabditis elegans'' ferlin, also known as ferlin-like proteins, therefore the name it was given, and can also be known as ferlin 1-like 1 (Fer1L1). It is common to this family the presence of type II transmembrane domains, where the most part of the protein faces de cytoplasm (3). This protein is critical for repair of muscle membranes after damage and its mutation lead to a progressive muscle dystrophy, since in its absence the membrane tear is not adequately repaired leading to myofiber necrosis and gradual and progressive loss of muscle tissue (1;5). The protein rapidly responds to injury with a calcium (Ca2+) influx mechanism which aids the repair. Dysferlin-deficient muscle fibers demonstrate a primary defect in Ca2+-dependent vesicle-mediated membrane repair (5). | + | <scene name='91/915204/Dysferlin/2'>Dysferlin</scene> is a large transmembrane protein of approximately 230kDa encoded by the dysferlin gene (DYSF omim) highly expressed in striated skeletal and cardiac muscle, but can be found in kidney, placenta, lung and brain tissues<ref [3]>. Dysferlin is a protein that belongs to the same family of genes as ''Caenorhabditis elegans'' ferlin, also known as ferlin-like proteins, therefore the name it was given, and can also be known as ferlin 1-like 1 (Fer1L1). It is common to this family the presence of type II transmembrane domains, where the most part of the protein faces de cytoplasm (3). This protein is critical for repair of muscle membranes after damage and its mutation lead to a progressive muscle dystrophy, since in its absence the membrane tear is not adequately repaired leading to myofiber necrosis and gradual and progressive loss of muscle tissue (1;5). The protein rapidly responds to injury with a calcium (Ca2+) influx mechanism which aids the repair. Dysferlin-deficient muscle fibers demonstrate a primary defect in Ca2+-dependent vesicle-mediated membrane repair (5). |
== Structure and Function == | == Structure and Function == | ||
Revision as of 15:18, 19 June 2022
Dysferlin
| |||||||||||
References
- ↑ . Dysferlin is a protein that belongs to the same family of genes as Caenorhabditis elegans ferlin, also known as ferlin-like proteins, therefore the name it was given, and can also be known as ferlin 1-like 1 (Fer1L1). It is common to this family the presence of type II transmembrane domains, where the most part of the protein faces de cytoplasm (3). This protein is critical for repair of muscle membranes after damage and its mutation lead to a progressive muscle dystrophy, since in its absence the membrane tear is not adequately repaired leading to myofiber necrosis and gradual and progressive loss of muscle tissue (1;5). The protein rapidly responds to injury with a calcium (Ca2+) influx mechanism which aids the repair. Dysferlin-deficient muscle fibers demonstrate a primary defect in Ca2+-dependent vesicle-mediated membrane repair (5).
Contents
Structure and Function
Structure
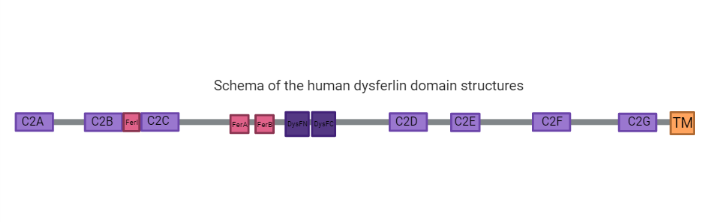
has seven tandem C2 domains (, B, C, D, E and G), three Fer domains (FerA, FerB and FerI) which are short conserved regions found only in the ferlin protein family and are not yet shown to be folded domains (2), two and a C-terminal transmembrane domain with a helix embedded in a patching vesicle.
C2 Domains
C2 domains are independently membrane-binding modules of about 130 residues found in a large and diverse set of eukaryotic proteins that share a common overall fold: a single compact greek-key motif organized as an eight-stranded antiparallel β-sandwich consisting of a pair of four-stranded β-sheets (6, 7). For example, its seen in the structure the C2A resolved to 2.04 Å by X-Ray diffraction the pair of four-stranded .
Fer Domains
Ferlins proteins are characterized by ferlin-specific small 60-70 residue motifs with conserved secondary structure termed FerI, FerA and FerB (6).
DysF Domain
One Dysf domain is inserted into the other Dysf domain forming an inner Dysf domain (not represented in the image below) and a two part outer Dysf domain (N-terminal DysFN and C-terminal DysFC), and it is woth mentioning that the Dysf domain is held together by arginine/ aromatic sidechain stacking. The crystal structure of the human dysferlin inner DysF domain with a resolution of 1.9 Ångstroms by X-Ray diffraction (2).
Function
Many dysferlinopathy causing mutations fall in the DysF domains (2). It's important to notice that dysferlin function is linked with calcium-activated membrane repair caused by fusing aggregated intracellular vesicles with the sarcolemma at the site of injury(2). It has been shown that dysferlin deficiency delays myoblast (undifferentiated mononuclear muscle cells) fusion/maturation in vitro, suggesting that dysferlin may also participate in muscle differentiation and regeneration process (3).
C2 Domains function
C2 domains are calcium sensitive phospholipid binding domains with an approximate length of 130 amino acids (5), while the function of the Dysf domain remains unclear <ref>DOI: https://doi.org/10.1371/journal.pone.0013854</li></ol></ref>
[3] https://www.omim.org/entry/603009?search=dysferlin&highlight=dysferlin; https://www.sciencedirect.com/science/article/pii/S0955067407000993