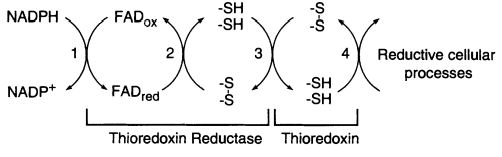
The of E. coli is a member of the flavoenzymes, responsible for catalyzing the reduction of the redox protein thioredoxin, by using NADPH. [1] The has its role as a protein disulfide reductase comprising thioredoxin reductase (TrxR), thioredoxin (Trx) and NADPH. Thioredoxin is the main disulfide reductase responsible for keeping intracellular proteins reduced. The redox reactions are catalyzed by thioredoxin reductase (TrxR) by taking electrons from NADPH and transferring them to the FAD domain of the oxidized Trx (Trs-S2). Electrons are then transferred to the redox active disulfide in the N-terminal of one subunit of TrxR, finally to the C-terminal active site of the other subunit, generating a a dithiol in reduced thioredoxin, Trx-(SH)2. [2] [3]
The activity of thioredoxin reductase on thioredoxin is importante in E. coli for more than a reason. Despite this enzyme has a conserved role as a high-capacity hydrogen donor systems for reductive enzymes, it also has evolved to specialized functions, such as participating in filamentous E. coli phage assembly and its T7 DNA replication. Furthermore, thioredoxin has demonstrated an important role in defense against oxidative stress or in control of apoptosis. Most of these functions are dependent of the disulfide reductase activity, which is only possible with the NADPH and thioredoxin reductase supply. [4]
Structure
The enzyme thioredoxin reductase from Escherichia coli is a homodimer, in which each monomer contains one and one redox active disulfide. The first part of the FAD binding domain (FAD-i), the NADPH binding domain, and the last part of the FAD binding domain (FAD-2) is formed by the polypeptide chain of each monomer. The connection between FAD and NADPH domains is a two-stranded, antiparallel B-sheet, and each one has a secondary and tertiary structure. [5]
The redox is formed by Cys135 and Cys138, located in the NADPH domain, adjacent to the re side of the flavin ring and interposed between the flavin
and NADPH binding site. [6] This structure conformation blocks the path of electrons from reduced pyridine nucleotide to the substrate disulfide via the isoalloxazine ring and the redox active disulfide, as it happens in the other member of this enzyme family. That distinction of the NADPH binding site from the flavin happened because of the juxtaposition of the FAD binding and NADPH binding domains, and it can be explained by conformational changes thioredoxin reductase suffer to enable the path of electrons in the catalyze. [7]
TrxR conformational changes
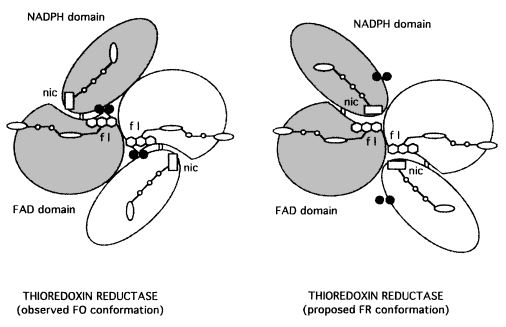
TrxR catalyzes the reaction of reduction of a protein substrate, thioredoxin, by NADPH. However, the enzyme structure does not reveal a clear path for reducing equivalents or has a binding site for thioredoxin. That paradox was solved by the suggestion of a large conformational change as a part of catalysis that generates the flavin-oxidizing (FO) and the flavin-reducing (FR) conformation of E. coli TrxR. [8]
In the FO conformation, the redox active disulfide present on the NADPH domain of TrxR is adjacent to the flavin ring in an orientation that allows electron transfer between them, reducing the disulfide with concomitant oxidation of the flavin. However, that orientation binds the active site disulfide blocking its interaction with the substrate, thioredoxin, preventing the protein reduction. [9] [10]
Thus, the FR conformation was modeled as a conformational change made by the enzyme to permit the active site disulfide reaction with the protein substrate. In this conformation, the NADPH domain rotates 67° about the axis toward the FAD resulting in the juxtaposition of the domains, exposing the disulfide loop from the interior of the enzyme and an important surface area for interaction with thioredoxin.[11] The position of NADPH also is favorable for the reduction of flavin by pyridine nucleotide by hydride transfer. This conformation permits the transference of reducing equivalents from reduced FAD to the active-site disulfide. That is possible because the dithiol is moved toward the solvent for interchange with the protein thioredoxin, and the pyridinium ring adjacent to the isoalloxazine ring for efficient hydride transfer.[12] [13]
References
- ↑ Williams CH Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995 Oct;9(13):1267-76. doi: 10.1096/fasebj.9.13.7557016. PMID:7557016 doi:http://dx.doi.org/10.1096/fasebj.9.13.7557016
- ↑ Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. 2010 May 21;396(1):120-4. doi:, 10.1016/j.bbrc.2010.03.083. PMID:20494123 doi:http://dx.doi.org/10.1016/j.bbrc.2010.03.083
- ↑ Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000 Oct;267(20):6102-9. PMID:11012661
- ↑ Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000 Oct;267(20):6102-9. PMID:11012661
- ↑ Williams CH Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995 Oct;9(13):1267-76. doi: 10.1096/fasebj.9.13.7557016. PMID:7557016 doi:http://dx.doi.org/10.1096/fasebj.9.13.7557016
- ↑ Williams CH Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995 Oct;9(13):1267-76. doi: 10.1096/fasebj.9.13.7557016. PMID:7557016 doi:http://dx.doi.org/10.1096/fasebj.9.13.7557016
- ↑ Lennon BW, Williams CH Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000 Aug 18;289(5482):1190-4. PMID:10947986
- ↑ Lennon BW, Williams CH Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000 Aug 18;289(5482):1190-4. PMID:10947986
- ↑ Lennon BW, Williams CH Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000 Aug 18;289(5482):1190-4. PMID:10947986
- ↑ Williams CH Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995 Oct;9(13):1267-76. doi: 10.1096/fasebj.9.13.7557016. PMID:7557016 doi:http://dx.doi.org/10.1096/fasebj.9.13.7557016
- ↑ Lennon BW, Williams CH Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000 Aug 18;289(5482):1190-4. PMID:10947986
- ↑ Lennon BW, Williams CH Jr, Ludwig ML. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci. 1999 Nov;8(11):2366-79. PMID:10595539
- ↑ Williams CH Jr. Mechanism and structure of thioredoxin reductase from Escherichia coli. FASEB J. 1995 Oct;9(13):1267-76. doi: 10.1096/fasebj.9.13.7557016. PMID:7557016 doi:http://dx.doi.org/10.1096/fasebj.9.13.7557016


