We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
FirstGlance/How To Measure A Virus Capsid
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
[[FirstGlance in Jmol]] automatically constructs the capsid, and simplifies it to a subset of alpha carbon atoms small enough (not more than 25,000) to be analyzed efficiently in FirstGlance and [[JSmol]], both of which run in the Javascript of the web browser. FirstGlance offers a number of useful color schemes, including distance from center, colors that distinguish sequence-identical groups of chains, and [[Help:Color_Keys#Rainbows:_N_to_C.2C_5.27_to_3.27|amino-to-carboxy rainbow]]. When the structure is small enough that all alpha carbons can be displayed (not more than 250,000 alpha carbons; EEEV has 242,340), it can also be colored by charge, hydrophobic vs. polar, or [[evolutionary conservation]]. | [[FirstGlance in Jmol]] automatically constructs the capsid, and simplifies it to a subset of alpha carbon atoms small enough (not more than 25,000) to be analyzed efficiently in FirstGlance and [[JSmol]], both of which run in the Javascript of the web browser. FirstGlance offers a number of useful color schemes, including distance from center, colors that distinguish sequence-identical groups of chains, and [[Help:Color_Keys#Rainbows:_N_to_C.2C_5.27_to_3.27|amino-to-carboxy rainbow]]. When the structure is small enough that all alpha carbons can be displayed (not more than 250,000 alpha carbons; EEEV has 242,340), it can also be colored by charge, hydrophobic vs. polar, or [[evolutionary conservation]]. | ||
| + | |||
| + | Quick Start: [http://bioinformatics.org/firstglance/fgij4/fg.htm?mol=6mx4 Analyze the EEEV capsid 6mx4 in FirstGlance]. | ||
| + | |||
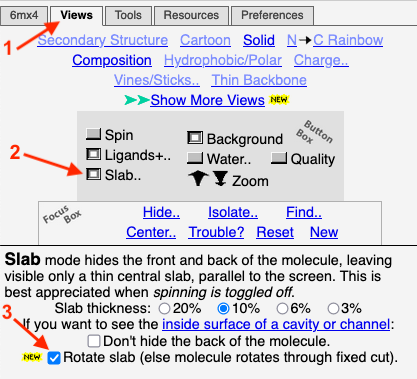
==Slab of the Capsid== | ==Slab of the Capsid== | ||
Revision as of 00:04, 28 July 2022
| |||||||||||