We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
FirstGlance/How To Measure A Virus Capsid
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
==Slab of the Capsid== | ==Slab of the Capsid== | ||
In order to estimate the dimensions of the capsid, it is useful to "cut out a slab". | In order to estimate the dimensions of the capsid, it is useful to "cut out a slab". | ||
| - | + | {{Template:EEEV-slab}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
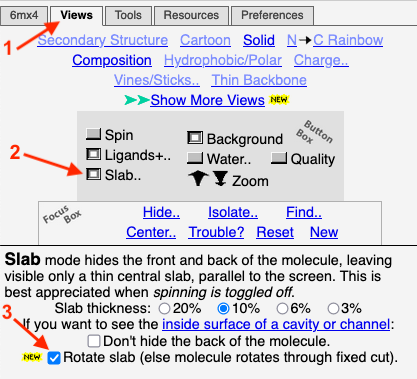
To obtain this slab in FirstGlance, you simply depress a ''Slab'' button. By default, the thickness of the slab is 10% of the diameter of the capsid (but this is adjustable). Here is a '''snapshot of the ''Views'' tab of FirstGlance'''. After the initial view of the capsid appears, getting this slab takes 3 clicks indicated by the red arrows: Click on the ''Views'' tab, depress the ''Slab'' button, and check ''Rotate slab''. | To obtain this slab in FirstGlance, you simply depress a ''Slab'' button. By default, the thickness of the slab is 10% of the diameter of the capsid (but this is adjustable). Here is a '''snapshot of the ''Views'' tab of FirstGlance'''. After the initial view of the capsid appears, getting this slab takes 3 clicks indicated by the red arrows: Click on the ''Views'' tab, depress the ''Slab'' button, and check ''Rotate slab''. | ||
Revision as of 00:07, 28 July 2022
| |||||||||||